
Special Article: Brain Cancer
Ann Hematol Onco. 2024; 11(1): 1447.
Epidemiology, Phylogeny and Drug Resistance Against Human Immuno- Deficiency Virus in Pakistan
Muhammad Basit Naeem¹; Junaid Ahmad¹; Bilal Arshad¹; Rubace Fatima Mirza¹; Rana Muhammad Sohail Afzalkhan¹; Rana Aetesam Nasir¹; Shaharyar Ashraf¹; Ifrah Saroosh¹; Muhammad Waqar Mazhar²*
1Department of Medicine and Surgery, Hitec-Institute of Medical Sciences Taxila Cantt, Pakistan
2Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan
*Corresponding author: Muhammad Waqar Mazhar Department of Bioinformatics and Biotechnology, Government College University, Faisalabad, Pakistan. Tel: +923012222861 Email: waqarmazhar63@gmail.com
Received: March 04, 2024 Accepted: April 09, 2024 Published: April 16, 2024
Abstract
Background: The human immunodeficiency virus is categorised in the genus Lentivirus, subfamily Orthoretrovirinae of the Retroviridae family.
Methodology: 40 specimens in total were included in the research to conduct the whole experimentation. The viral RNA extraction was performed by using the Qiagen viral RNA extraction kit. After the extraction of RNA, the cDNA was synthesized. Qiagen One step rt-PCR kit was used for amplification by using pair of primers forward primers (A) & reverse primer (D) 10uM/ug. Sequencing and capillary electrophoresis were performed. The phylogenetic tree of HIV was done by using mega.
Results: Out of 40, 10 samples are sequenced 4 of them are resistance against drugs. Sample Ids 226 and 229 are resistance to Rilpivirine (RPV) and Etravirine (ETR) also has 138A mutation inHIV-1. Sample Id 231 have resistance to Lamivudine (3TC), Emtricitabine (FTC) and Abacavir (ABC) also has M184V mutation in HIV-1. Sample Id 232 have resistance to Lamivudine (3TC), Emtricitabine (FTC) and Abacavir (ABC) also has M184V Nevirapine (NVP) and Efavirenz (EFV) mutation in HIV-1.
Keywords: HIV; Retroviridae; Phylogenetic tree; Drug Resistance Mutation
Introduction
The virus known as Human Immunodeficiency Virus (HIV) targets immune system cells, increasing a person's susceptibility tovarious maladies and diseases [18,22]. The Human Immunodeficiency Virus (HIV) is a member of the retroviral family Orthoretrovirinae and belongs to the genus Lentivirus [19]. HIV is divided into types 1 and 2 according to genetic attributes and variations in the viral proteins (HIV-1, HIV-2). HIV- 1 is divided into three classes: group M (main), group O (outlier), and group N [8]. The far more varied group, Group M, is made up of nine subtypes (A-D, F-H, J, and K) and many circulating recombinant forms (CRFs) [11,24]. The prevalence of the subtypes geographically and demographically is varied, including one or more variants dominate infection in certain geographical regions; for instance, subtype C predominates in Southern and Eastern Africa, as well as China. [7,10]. The incidence of non-B subtypes and infection is expected to rise in the industrialized western world, along with the US, because of increased rates of mobility and interaction with people from non-B endemic areas [23]. Regarding this tendency, most research-based efforts have been focused on the geographically prevalent subtype B in North America and Europe [25] According to recent studies, subtypes C and A together account for more than 70% of all new infections, whereas HIV-1 subtype B only accounts for 12% of the approximated 40 million HIV- infected people globally [28].
The prevalence and epidemiology of HIV in Pakistan, when compared to other nations in the region, is comparatively low. According to the Joint United Nations Program on HIV/AIDS (UNAIDS), the approximated number of HIV-positive individuals in Pakistan is 150,000 as of 2019, representing around 0.1% of the population [21]. According to estimates, 183,705 persons in Pakistan are HIV positive (PLHIV). Individuals who inject drugs (PWID), male, female, and transgender sex workers (MSW, FSW, & TGSW), men who have sex with men (MSM), and transgenders are among the primary demographics where the HIV pandemic in the nation is centered [2]. HIV can cause AIDS if not properly treated by sharing injecting equipment or coming into touch with certain bodily fluids of an HIV-positive person, most commonly during unprotected sexual activity. (Anwar et al.) Antiretroviral Treatment (ART) advancements have led to notable successes, among them the significant reductions in morbidity and death seen in HIV-infected individuals. The virus's main survival strategy, which derives from its enormous ability to produce variation, is Antiretroviral (ARV) medication or drug resistance [14,21] An evaluation of 117 sources has provided information on the general frequency of ARV resistance in the developing world, with an emphasis on treatment-naive individuals, the effects of Prevention of mother-To-child Transmission (PMTCT) drug regimens on resistance, and the connection between medication adherence and resistance [9]. Global treatment-naive populations' patterns of ARV resistance seem to match regional patterns in the use of ARV drugs. The frequency of resistance (to any medicine) among people who have never had treatment was reported to be 5.5% in Africa, 7.4% in East Asia, 5.7% in Southeast Asia, and 6.4% in Latin America, lower than the rates in North America (11.4%) and Europe (10.6%) [1].
Pre-exposure prophylaxis, a medicine that people who are at risk for catching HIV use to prevent contracting HIV through sex or injecting drugs, is one of the efficient ways to prevent contracting HIV through sex or drug use, and Post-Exposure Prophylaxis (PEP), a medication that HIV-positive patients must take within 72 hours of an increased risk to cease the virus from taking hold. Learn more about additional HIV prevention strategies [3,4].
Materials & Methods
Statement
No cross-sectional survey of HIV-1 has been done to date to provide a true depiction of the epidemiology in the nation especially targeting Punjab because it is needed to develop better public health policies related to the prevention and therapies, despite the fact that it may seem exhausting to invest in a disease that typically presents its fate, improperly tapping into the source of the disease can lead to its spread to high-risk groups. Additionally, it is necessary to alleviate the disease's morbidities, and genotyping can show which principal vector the virus used to propagate.
Ethics Statement Study Group
40 specimens in total were included in the research to conduct the whole experimentation. This number of 40 totally satisfied the calculation for the cross-sectional epidemiological study of HIV-1.
Z= Zē.p(1-p)/dē
Whereas,
Z=Standard normal variate (for 5% error =1.96, (p<0.05)),
p= expected proportion of population’s disease in previous research; In Pakistan for HIV-1 it’s 0.01
d= absolute error/precision
If we put the values in the given formula with 5% error rate (p<0.05), we got 12 & we got higher number of samples than this to strengthen this study. To conduct this cross-sectional study a certain group of patients comprised in this research, All the patients were HIV-1 positive and got their treatment from the Punjab AIDS control program, inclusion criteria is simple as this study mainly focus on DRMs, so this study includes that patients of HIV-1 who were on the treatment of HIV-1. Sanger sequencing done for the identification of DRMs in HIV-1 positive samples, following steps done for this experimentation.
RNA Extraction and cDNA Synthesis
Collect the serum sample and centrifuge it to pellet the cells. The viral RNA extraction was performed by using the Qiagen viral RNA extraction kit (catalog no. 52904). After the extraction of RNA from samples cDNA synthesized with the reverse primer (D). For this experimentation many primers were used for the maximum coverage of the pol region. A list of primers was given in the given table which were used in this experimentation. For this purpose, Qiagen One step rt-PCR kit used where cDNA and desired Round 1 product is being amplified by using pair of primers forward primer (A) & reverse primer (D) 10uM/ug.
Virus
Primer name
Primer Sequence 5’ to 3’
Region in pol
Direction
HIV-1
A
CAGGAGCAGATGATACAG
Protease
Forward
Bf
TGGACTGTCAATGATATACA
Reverse Transcriptase
Forward
Br
TGTATATCATTGACAGTCCA
Reverse Transcriptase
Reverse
CF
ACAGTGCAGGGGAAAGAA
Integrase
Forward
Cr
TTCTTTCCCCTGCACTGT
Integrase
Reverse
D
CCCTTCACCTTTCCAGAG
Integrase
Reverse
a1
ATAGGGGGAATTGGAGGTTTTAT
Protease
Forward
a2
AGGAATGGATGGCCCAAA
Protease
Forward
b1
GGGTTATGAACTCCATCCTGATAAATGGAC
Reverse Transcriptase
Forward
b2f
TGGAGAGCAATGGCTAGTGA
Reverse Transcriptase
Forward
b2r
TCACTAGCCATTGCTCTCCA
Reverse Transcriptase
Reverse
Table 1: Shows the primers used in this experimentation (cDNA synthesis, round 1 & 2 PCR, and cycle sequencing PCR). All these 11 primers are used to amplify the maximum region of pol region in HIV-1 which is the most vulnerable region for DRMs.
Cycle Sequencing PCR
As the PCR products were purified by using HighPrep PCR clean up, These products served as the cycle sequencing PCR template. For each PCR product a specific combination of primers was used for sequence that region. The following table shows the primers which were used to sequence that region (Table 2).
Sr. No.
Primers Combination for PCR
Primers used to sequence for that region
1
A-Br
A, a1, a2, Br
2
Bf-Cr
Bf, b2r, b2f, Cr
3
Cf-D
Cf & D
4
a2-b2r
a2, b1, Br, Bf, b2r
5
b2f-D
b2f, Cr, Cf, D
6
b1-Cr
b1, Bf, b2r, b2f, Cr
Table 2:
Capillary Electrophoresis
Purified cycle sequencing product was now ready for capillary electrophoresis, where in the end extracted sequence can be seen in the form of chromatograms. 40 ul of purified product poured in the 96-well plate &sealed with septa. The plate was now ready for the 3500XL Genetic Analyzer which was used for this procedure.
Data Analysis Sequence editing
Sequencing files produced by 3500xl in the form of ABI files. This is the raw data which was trimmed by the Sequencher software to make a final FASTA file.
• Launch Sequencher and open the ABI files by going to File > Open > ABI File.
• Select the ABI files that you want to edit and click Open.
• The ABI files will be displayed in the Sequencher window.
• Use the trimming and editing tools in Sequencher to remove any low-quality or incorrect bases from the sequences. You can use the Trim tool to remove bases from the ends of sequences, or the Edit tool to make changes to the sequences.
• Once you have finished editing the sequences, you can export them as a consensus FASTA file by going to File > Export > Consensus FASTA.
• In the Export Consensus FASTA dialog box, select the sequences that you want to include in the FASTA file, and state the output file name and location.
For the construction of phylogenetic tree MEGA software used https://megasoftware.net/.
Detection of Drug Resistance Mutations (DRMs)
DRMs can be find by submitting sequences (FASTA file) in the “Stanford University HIV Drug resistance Database” https:// hivdb.stanford.edu/. Where it’s not only detects the DRMs but also characterize into its subtype & also finds the drugs that are resistance to the HIV virus.
Results
Gel Electrophoresis
For tracking the PCR progress agarose gel was made for the visualizing of amplicons through gel electrophoresis experiment. 1.5% agarose gel made for this experiment, and it run on the Voltage of 110 for 45 minutes. To judge the exact size of amplicon 100bp Ladder run as a control.
Sequence Alignment & Phylogenetic Tree
Sequences of additional HIV-1 subtypes were acquired in FASTA format from NCBI in order to evaluate the phylogenetic context of the sequenced samples. The additional sequences which were extracted from NCBI were: KC203330.1-KC203332.1 (China), KT581450.1 (Germany), JX227940.1 (Senegal), JX227940.1, KF890251.1, KF890250.1 (India), MT240851.1, MT240850.1, MT223668.1, MT223667.1, MT223665.1 (Pakistan), AY010480.1, FM164924.1 HIV-1 (France), L23102.1, FM164931.1 (Taiwan).
These all sequences include different subtypes which make a tree to validate previous study and it also strengthens this study. A rooted tree construct for this study with HBX2 as a reference sequence.
Detection of Drug Resistance Mutations (DRMs)
The Stanford HIV Drug Resistance Database (HIVDB) is an important tool for public health workers who are keeping track of ADR and TDR, scientists who are making new ARV drugs, and HIV care providers who are taking care of HIVDR patients. HIVDR researchers also use HIVDB to compare their findings to those of other studies and to do meta-analyses, which require data from many studies and make it possible to learn new things that can't be learned from just one survey.
To summarize the outcome in sequenced samples that makes a relation between drug resistance mutations with drugs can be seen in the gplot given below.
Interpretation of Above Plot
In the above plot out of 10 sequenced samples 4 samples were resistance towards the following drugs as shown in the y-axis of the graph. All the drugs are the inhibitors for reverse transcriptase region in HIV-1. Relation of each mutation with its drug described below. Sample Ids 226 and 229 are resistance to Rilpivirine (RPV) and Etravirine (ETR) also has 138A mutation inHIV-1. Sample Id 231 have resistance to Lamivudine (3TC), emtricitabine (FTC) and abacavir (ABC) also has M184V mutation in HIV-1. Sample Id 232 have resistance to Lamivudine (3TC), emtricitabine (FTC) and abacavir (ABC) also has M184V Nevirapine (NVP) and Efavirenz (EFV) mutation in HIV-1.
Discussion
In this century the main problem is drug resistance in viruses. HIV known as fast evolving virus, treatment was only effective for a few months because of drug resistance mechanism [5]. The Present study investigated the drug resistance against HIV-1 in pakistan. These strains might enter the population as a result of the emigration of HIV-positive people, such as labourers and sex workers, from countries including India, Iran, and Afghanistan [29]. The Middle East, and Africa. 2017's analysis of the virus's geographic distribution in Pakistan revealed that sequences entered into the database up until that point were tightly associated with sequences from Afghanistan, India, South Africa, Kenya, and Rwanda [29]. Our findings were in line with this, showing that sequences were grouping with nations like Yemen, South Africa, and Uganda in the Middle East, as well as with African nations like Uganda and South Africa (Klotz, 2022) However, 18.75% of the sequences are new subtypes and are grouped with Tanzania, Afghanistan, and Sweden. These connections between HIV strains that are proliferating in other nations might be the result of infected people entering and leaving Pakistan without a medical clearance or surveillance. As a result, it is necessary to update immigration regulations in order to stop future contagious spread. Drug resistance is rising along with the trend of genetic variation, and the virus is developing high-level resistance due to vulnerability to ART medications.
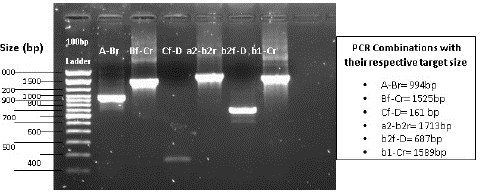
Figure 1: Gel Electrophoresis picture shows the amplified product of HIV-1 pol’s region.
Figure 2: Sequencher 4.9 dashboard | Chromatograms for the editing of sample.
Figure 3: Primers overlapped in Sequencher 4.9 software covers the pol region of HIV-1.
Figure 4: Alignment of sequenced samples and retrieved sequences from NCBI database.

Figure 5: A Phylogenetic tree of 28 representative sequences of human immunodeficiency virus (HIV). This tree was generated using Neighbor Joining method in MEGAX. HXB2 using as the reference sequence, detected isolates of HIV are identified with red circle node markers (.). The distance scale represents the number of differences between the sequences.
Figure 6: Stanford University HIV Drug resistance Database Dashboard.
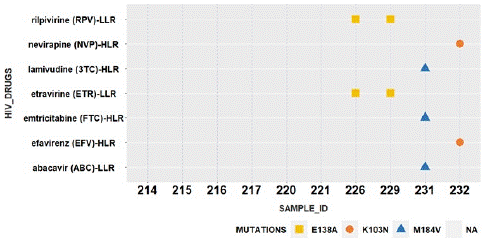
Figure 7: A R-Plot shows the relation of 10 sequenced samples (x-axis) with drugs (y-axis) used against the HIV-1 Infected patients with their specific related mutation.
The RPV and ETR are Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) that are utilized to treat HIV-1. The E138A mutation in HIV-1 is a resistance-associated mutation that can occur in the Reverse Transcriptase (RT) enzyme of the virus. (Himmel & Arnold, 2020; Wang, De Clercq, & Li, 2019) This mutation can confer resistance to NNRTIs like RPV and ETR. This means that if a person's HIV-1 strain has the E138A mutation, it may be less susceptible to the antiviral effects of RPV and ETR, making these drugs less effective in treating the virus.
3TC, FTC and ABC are Nucleoside Reverse Transcriptase Inhibitors (NRTIs) that are utilized to treat HIV-1. The M184V mutation in HIV-1 is a resistance-associated mutation that can occur in the reverse transcriptase (RT) enzyme of the virus. (Cilento et al., 2021; Lai et al., 2022; Marin, Streinu-Cercel, Moleriu, & Bungau, 2022) This mutation can confer resistance to NRTIs like 3TC, FTC and ABC, meaning that if a person's HIV-1 strain has the M184V mutation, it may be less susceptible to the antiviral effects of these drugs, making them less effective in treating the virus. This is one of the most common mutations that happen in HIV-1 and is considered as a significant genetic marker for NRTI resistance.
NVP and EFV are Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs) that are utilized to treat HIV-1. The K103N mutation in HIV-1 is a resistance-associated mutation that can occur in the Reverse Transcriptase (RT) enzyme of the virus. (Huang et al., 2019; Wang et al., 2019; Z. Wang et al., 2022). This mutation can confer resistance to NNRTIs like NVP and EFV, meaning that if a person's HIV-1 strain has the K103N mutation, it may be less susceptible to the antiviral effects of these drugs, making them less effective in treating the virus. The K103N mutation is considered as among most common NNRTI resistance mutations and it is often associated with cross-resistance to other NNRTIs.
Drug resistance for HIV has not yet been resolved. In actuality, patients with coexisting conditions are more likely to have treatment resistance. For instance, a pregnant HIV-positive woman who lacks access to appropriate care might receive therapy with sdNVP to stop mother-to-child transmission. (Nguyen, Ton, Tran, & Nguyen, 2020) Because sdNVP use can result in NNRTI resistance, if she later receives regular medication, it could not be effective. Additionally, if she stays on the ineffective treatment and picks up more drug-resistance mutations, which can jeo pardise second-line therapies, and if she doesn't live in a region where viral load monitoring is possible. Drugs and monitoring need to be much more widely available in order to address the issue of drug resistance.
Conclusion
These connections between HIV strains that are proliferating in other nations might be the result of infected people entering and leaving Pakistan without a medical clearance or surveillance. As a result, it is necessary to update immigration regulations in order to stop future contagious spread. Drug resistance is rising along with the trend of genetic variation, and the virus is developing high-level resistance due to vulnerability to ART medications.
Author Statements
Data Availability Statement
The data generated during this study has been included in the manuscript.
Conflicts of Interest
“The authors declare no conflict of interest.”
Funding
No external funding was received.
References
- Abubakari A, Issah H, Mutaka M, Asumah, MN. Determinants of Virological Failure in HIV Patients on Highly Active Antiretroviral Therapy (HAART): A Retrospective Cross-Sectional Study in the Upper East Region of Ghana. Venereology. 2023; 2: 16-29.
- Anwar A, Ali H, Qamar F, Abbas S, Jamil B, Akhtar N. HIV Testing Care and Treatment: A Guide for Health Care Providers in Pakistan. 2023: 4.
- Auerbach JD, Kinsky S, Brown G, Charles V. Knowledge, attitudes, and likelihood of pre- exposure prophylaxis (PrEP) use among US women at risk of acquiring HIV. AIDS patient care and STDs. 2015; 29: 102-110.
- Bazzi AR, Biancarelli DL, Childs E, Drainoni ML, Edeza A, Salhaney P, et al. Limited knowledge and mixed interest in pre-exposure prophylaxis for HIV prevention among people who inject drugs. AIDS patient care and STDs. 2018; 32: 529-537.
- Bloch M, John M, Smith D, Rasmussen T, Wright E. Managing HIV-associated inflammation and ageing in the era of modern ART. HIV medicine. 2020; 21: 2-16.
- Cilento ME, Reeve AB, Michailidis E, Ilina TV, Nagy E, Mitsuya H, et al. Development of Human Immunodeficiency Virus Type 1 Resistance to 4'-Ethynyl-2-Fluoro-2'- Deoxyadenosine Starting with Wild-Type or Nucleoside Reverse Transcriptase Inhibitor-Resistant Strains. Antimicrobial Agents and Chemotherapy. 2021; 65: e01167-01121.
- Daw MA, El-Bouzedi AA, Ahmed MO, Dau AA, Agnan MM, Drah AM. Geographic integration of hepatitis C virus: a global threat. World journal of virology. 2016; 5: 170-182.
- Dubey A. Machine learning models for evaluation of domain Based classification of HIV-1 groups. Online Journal of Bioinformatics. 2017; 18: 53-57.
- Hamers RL, Derdelinckx I, van Vugt M, Stevens W, Rinke de Wit TF, Schuurman R. The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antiviral therapy. 2008; 13: 625-639.
- Han X, Xu P, Wang H, Mao J, Ye Q. Incident changes in the prevalence of respiratory virus among children during COVID-19 pandemic in Hangzhou, China. Journal of Infection. 2022; 84: 579- 613.
- He L, Dong R, He RL, Yau SST. A novel alignment-free method for HIV-1 subtype classification. Infection, Genetics and Evolution. 2020; 77: 104080.
- Himmel DM, Arnold E. Non-nucleoside reverse transcriptase inhibitors join forces with integrase inhibitors to combat HIV. Pharmaceuticals. 2020; 13: 122.
- Huang B, Chen W, Zhao T, Li Z, Jiang X, Ginex T, et al. Exploiting the tolerant region I of the non-nucleoside reverse transcriptase inhibitor (NNRTI) binding pocket: discovery of potent diarylpyrimidine-typed HIV-1 NNRTIs against wild-type and E138K mutant virus with significantly improved water solubility and favorable safety profiles. Journal of Medicinal Chemistry. 2019; 62: 2083-2098.
- Kharsany AB, Karim QA. HIV infection and AIDS in sub-Saharan Africa: current status, challenges and opportunities. The open AIDS journal. 2016; 10: 34.
- Klotz A. The Southern Africa Migration System. Understanding Global Migration. 2022.
- Lai MT, Feng M, Xu M, Ngo W, Diamond TL, Hwang C, et al. Doravirine and Islatravir Have Complementary Resistance Profiles and Create a Combination with a High Barrier to Resistance. Antimicrobial Agents and Chemotherapy. 2022; 66: e02223-02221.
- Marin RC, Streinu-Cercel A, Moleriu LC, Bungau SG. Analysis of virological response to therapy and resistance profile in treatment-experienced and naive HIV-1 infected Romanian patients receiving regimens containing darunavir boosted with ritonavir or cobicistat. Biomedicine & Pharmacotherapy. 2022; 150: 113077.
- Mehmood J, Saif S, Iram W, Javeed H, Iftikhar H, Khan U, et al. Corelation of HIV With Prostitution In Punjab. Asian Journal of Advances in Medical Science. 2021; 187-189.
- Mozhi PK, Ganapathy D. Awareness of Structural Biology of HIV Among Dental Students. European Journal of Molecular and Clinical Medicine. 2021; 8: 491-504.
- Nguyen RN, Ton QC, Tran QH, Nguyen TKL. Mother-to-child transmission of HIV and its predictors among HIV-exposed infants at an outpatient clinic for HIV/AIDS in Vietnam. HIV/AIDS-Research and Palliative Care. 2020; 12: 253-261.
- Paul D, Rodes B, Cadinanos J, Esteban-Cantos A, Rodríguez-Centeno J, Arribas JR. The comprehensive review of antiretroviral therapy: a paradigm shift. Brac University. Ageing with HIV: Challenges and biomarkers. EBioMedicine. 2022; 77: 103896.
- Sillman B, Woldstad C, Mcmillan J, Gendelman HE. Neuropathogenesis of human immunodeficiency virus infection. Handbook of Clinical Neurology. 2018; 152: 21-40.
- Stella L, Santopaolo F, Gasbarrini A, Pompili M, Ponziani FR. Viral hepatitis and hepatocellular carcinoma: From molecular pathways to the role of clinical surveillance and antiviral treatment. World Journal of Gastroenterology. 2022; 28: 2251.
- Tebit DM, Nankya I, Arts EJ, Gao Y. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle. Aids Rev. 2007; 9: 75-87.
- Wang SS, Vajdic CM, Linet MS, Slager SL, Voutsinas J, Nieters A, et al. B-Cell NHL Subtype Risk Associated with Autoimmune Conditions and PRS. Cancer Epidemiology, Biomarkers & Prevention. 2022; 31: 1103-1110.
- Wang Y, De Clercq E, Li G. Current and emerging non-nucleoside reverse transcriptase inhibitors (NNRTIs) for HIV-1 treatment. Expert opinion on drug metabolism & toxicology. 2019; 15: 813-829.
- Wang Z, Cherukupalli S, Xie M, Wang W, Jiang X, Jia R, et al. Contemporary medicinal chemistry strategies for the discovery and development of novel HIV-1 non- nucleoside reverse transcriptase inhibitors. Journal of Medicinal Chemistry. 2022; 65: 3729-3757.
- Yuan D, Zhong X, Li Y, He Q, Li N, Li H, et al. Molecular Transmission Network of Newly Reported HIV Infections in Pengzhou, Sichuan Province: A Study Based on Genomics and Spatial Epidemiology. International Journal of Environmental Research and Public Health. 2023; 20: 2523.
- Zahid M, Khan S, Qureshi MA, Raza Y. Subgenomic sequence analysis reveals emergence of new circulating recombinant forms of HIV-1 in Pakistan. Pakistan Journal of Medical Sciences. 2022; 38: 1857-1863.