
Editorial
Ann Hematol Oncol. 2015; 2(7): 1054.
Tracking Down the Origin of Stem Cell Programs in Cancer Cells
Ke F¹, Cai YJ¹, Tang JY² and Hong DL1,2
¹Key Laboratory of Cell Differentiation
and Apoptosis of National Ministry of Education,
Department of Pathophysiology, Shanghai Jiao Tong
University School of Medicine, Shanghai, China ²Key Laboratory of Pediatric Hematology & Oncology
Ministry of Health, Department of Pediatric Hematology
and Oncology, Shanghai Children’s Medical Center,
Shanghai Jiao Tong University School of Medicine,
Shanghai, China *Corresponding author: Hong DL, Key Laboratory of
Cell Differentiation and Apoptosis of National Ministry
of Education, Department of Pathophysiology, Shanghai
Jiao Tong University School of Medicine, 280 South
Chongqing Road, 200025, Shanghai, China Received: September 06, 2015; Accepted: October 15, 2015; Published: October 17, 2015 Cancer stem cells (CSCs) are cells that possess stem cell properties,
particularly the ability to propagate cancer. The origin of the stem cell
program of cancer cells is a key issue elucidating carcinogenesis. It is
possibly retained from stem cells or cancerously reprogrammed from
progenitor and even mature cells. Studies on leukemia have provided
pivotal observations, particularly the identification of initiating premalignant
cells, namely, pre-leukemic stem cells (pre-LSCs) [1] or
pre-leukemic hematopoietic stem cells (HSCs) in leukemogenesis [2]. Initiating pre-malignant cells are difficult to identify because of
lack of feasible approaches to determine healthy individuals who
carried the cells. Genetic analysis has provided opportunities in this
endeavor. For the first time, we identified initiating pre-malignant
stem cells in leukemia by using the unique genetic background of a pair
of monochorionic twins [1]. In our study, a 2-year-old twin acquires
acute lymphoblastic leukemia (ALL), whereas the other twin remains
healthy. Molecular analysis of the blood cells of the twins revealed
the existence of an identical leukemic fusion, namely, TEL-AML1.
The fusion was considered prenatal in origin, that is, it occurred in
utero in one twin and then spread to the other twin through their
shared placenta. TEL-AML1 has been presumed as the initiating
genetic lesion in associated leukemia [3]. Immunophenotypic
analysis of blood cells of the healthy twin revealed a population
containing markers for stem cells (CD34+ and CD38−) and B-cells
(CD19+). This population does not exist in normal blood samples.
Subsequently, the population was replicated in our disease models by
inducing TEL-AML1 in human cord blood (CB) cells, followed by
xenotransplantation in NOD-SCID mice or inoculation in cultures
with mouse stromal cells, MS5. The results showed the self-renewal
potential of the population in both experimental systems and thus
demonstrated the population contained pre-LSCs [1]. Modern advanced genetic techniques, including deep sequencing,
have been used to determine founder mutations that may generate
a pre-malignant ancestral cell of cancer. Pre-leukemic HSCs have
been recently identified in acute myeloid leukemia (AML) after the
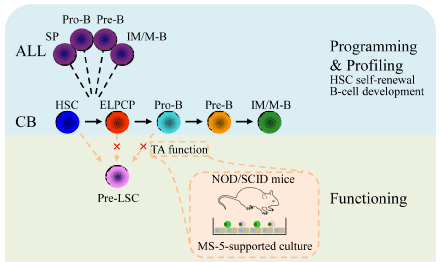
initiating mutation DNMT3Amut was determined [2]. We investigated the early reprogramming process of
leukemogenesis by obtaining samples from pre-malignant cells and
subjecting them to RNA array analysis. We determined that modeled
pre-LSCs were located at the differentiation stage between HSCs
(CD19−, CD34+, and CD38−) and pro-B cells (CD34+ and CD19+).
Functional assay results showed that pre-LSCs were primed with
multi-lineage potential; hence, stem cell programs in pre-LSCs may
be retained from HSCs, rather than converted from B progenitor
cells. This assumption was proven by the results of leukemogenesis
targeting experiments, in which only cells that possess self-renewal
potential (that is, HSCs) are the suitable target of TEL-AML1 (Figure
1) [4]. To consolidate previous findings, we investigated samples
of patients with TEL-AML1-associated ALL; these samples were
functionally confirmed to have stem cell properties and contain
leukemic stem cells (LSCs) within different immunophenotypic
fractions. We comparatively profiled a refined program in cells of
the immunophenotypic populations of human ALL and CB cells of
normal counterparts at different differentiation stages; this program,
which was termed as HSCB program, was edited from functional genes
essential to HSC self-renewal and B-cell development. Bioinformatic
analysis showed that ALL populations were loosely clustered and
located close to the normal population, which contains stem and
primitive progenitor cells (Figure 1). This result confirms that stem
cell programs in leukemic lymphoblast’s are retained from stem
cells, rather than introduced on progenitor cells by leukemogenic
molecules, such as TEL-AML1 [4]. Methylation analysis of self-renewal genes was conducted to
investigate the mechanism underlying the retention of stem cell
programs in pre-LSCs and LSCs. The results showed that these genes
are active and exhibit a low methylation level in HSCs and become
hypermethylated and inactive in progenitor cells; by contrast, these
genes consistently present a low methylation level in leukemic cells
with stem cell properties. Furthermore, this maintenance of the
stem cell program may occur in specific niches because TEL-AML1-
expressed cells tend to localize closely along the endosteum in bone
marrow (BM) (unpublished data) [5]. The machinery of self-renewal differs between fetal HSCs and
BM HSCs, and childhood leukemia may have a prenatal or postnatal
origin [6]. Future studies must investigate the mechanism underlying
the conversion of fetal self-renewal machinery to a postnatal one when
HSCs migrate to BM after birth. Findings of such studies are crucial
in elucidating the mechanism through which stem cell programs are
cancerously reprogrammed to trace down the origin of leukemia.
Factors such as niche adaption, epigenetic alteration, and genetic
mutation may be mutually involved in the process [5]. The developed concept and experimental paradigm used may be applicable for
exploring similar issues in other types of cancers in various tissues.
Editorial
References