
Review Article
Ann Hematol Oncol. 2022; 9(4): 1405.
Genomic Alterations in Acute Myeloid Leukemia in Routine Clinical Practice
Benková K1,2, Jelínek T1,2, Hájek R1,2 and Korístek Z1,2*
1Department of Hematooncology, University Hospital Ostrava, Ostrava, Czech Republic
2Department of Hematooncology, Faculty of Medicine, University of Ostrava, Ostrava, Czech Republic
*Corresponding author: Zdenek Korístek, Department of Hematooncology, Faculty of Medicine, University Hospital Ostrava, 17. listopadu 1790/5, Ostrava 708 52, Czech Republic
Received: August 17, 2022; Accepted: September 16, 2022; Published: September 23, 2022
Abstract
Acute Myeloid Leukemia (AML), the most common acute leukemia in adults, is a genetically heterogeneous disease. Genomic alterations condition the pathophysiology of AML. Nowadays, these changes are considered to be important biomarkers for risk stratification, treatment decisions (including new targeted drugs) and Minimal Residual Disease (MRD) monitoring during followup of AML. Positive MRD in AML patients is associated with a higher risk of relapse and shorter overall survival compared to MRD negative individuals. Nevertheless, MRD-targets, diagnostic techniques, time points of MRD determination, and analyzed material are not standardized. Thus, integration of MRD testing in individual AML patients in routine practice is still a workin- progress. This review article comprehensively focuses on key molecular biomarkers in AML and their utilization in clinical practice, especially regarding risk assessment and MRD testing. Moreover, new AML molecular-targeted therapies are briefly summarized here.
Keywords: Acute myeloid leukemia; Genomic alterations; Molecular biomarkers; Minimal residual disease; Targeted therapies
Introduction
Acute myeloid leukemia is characterized by uncontrolled proliferation and accumulation of clonal immature myeloid cells in bone marrow and peripheral blood leading to hematopoietic failure [4]. AML is a genetically heterogeneous disease with a very variable prognosis and high mortality rate. The 5-years Overall Survival (OS) is less than 50% and only 20% of elderly patients will survive 2 years after diagnosis [48]. Nowadays, cytogenetic profile and new molecular markers play an important role in diagnosis, prognosis, and monitoring of AML, as well as in optimizing therapeutic strategies [48].
Achieving complete remission defined by the absence of leukemic blasts in the bone marrow is not sufficient for predicting longterm remission as most patients relapse [63]. This prediction was improved with measuring the residual levels of leukemic cells (also called minimal or measurable residual disease; MRD) that persist in the bone marrow after chemotherapy [41]. In current studies, MRDtargets, diagnostic techniques, time points of MRD assessment, and analyzed material are variable. With the exception of MRD detection in Acute Promyelocytic Leukemia (APL), for which standardized guidelines have been published [54], there is still a lack of prospective randomized studies providing data on how to modulate treatment to alter outcomes based on MRD status in non-APL AML. It has been proven that MRD assessment allows outcome prediction. So far, implication of MRD testing in individual AML patients in routine practice is still a work-in-progress [28].
In this work, we comprehensively review key molecular biomarkers in AML, focusing on their utilization in clinical practice. We emphasize risk stratification and MRD testing, as well as novel AML targeted therapy. Routinely used methods to determine MRD and some future perspectives, and finally, practical aspects of MRD monitoring are also briefly presented.
Techniques for MRD Assessment
In practice, MRD evaluationis commonly performed by two methods – Multicolor Flow Cytometry (MFC) and real time quantitative PCR (RT-qPCR). Modern techniques such as Next Generation Sequencing (NGS) and digital droplet PCR (ddPCR) could be applied to routine clinical practice in the near future on account of their benefits [28].
Multicolor Flow Cytometry
Multicolor flow cytometry is a technique recognizing antigens on leukemic cells by fluorescence-labeled antibodies [28]. MRD detection by MFC is rapid and applicable to almost all AML patients (up to 90%), but has lower sensitivity (10-3–10-4) when compared to other techniques [57,67]. Two partially overlapping analyzing strategies can be used for MFC-MRD; the first focuses on Leukemia- Associated Immunophenotypes (LAIP), which are determined at the time of diagnosis and then used to track down residual leukemic cells in the follow-up samples. The second one is based on identifying any immunophenotypes that are Different-from-Normal (DfN) in samples submitted for MRD analysis [18,28,57,67]. Currently, the integrated LAIP-based DfN approach is recommended by the European Leukemia Net (ELN) MRD Working Party [56]. Unfortunately, MRD monitoring by MFC has not been fully standardized in AML in comparison to other hematologic malignancies [27].
PCR-Based Methods
Polymerase Chain Reaction (PCR) is a method that allows quantification of nucleic acids by amplification with the enzyme DNA polymerase [8]. In AML, these techniques identify AML-associated mutations in the follow-up samples. Real-time quantitative PCR is the golden standard for molecular MRD detection. RT-qPCR provides high sensitivity (10-4–10-6) and specificity, however, it can be used only in those patients harbouring specific aberrations (especially NPM1, CBF-MYH11, RUNX1-RUNX1T1, PML-RARA, accounting for approximately 40% of all AML patients) [18].
An innovative method called droplet digital PCR provides high precision quantification which is very reliable and offers the possibility to monitor several mutations simultaneously. However, it is not currently being used in routine practice, mainly due to higher costs and a limited number of equipped laboratories [8,18,28].
Next Generation Sequencing
NGS allows us to increase our knowledge of molecular heterogeneity in AML. Due to broader mutation coverage, it has the potential to be used in almost all AML patients. However, the sensitivity of NGS assays depends on DNA quality and quantity, as well as on the distinct monitored mutations. Universally standard quality criteria for NGS have still not been determined. Therefore, its application in routine practice has to be individually validated [1,28,57,66].
Molecular Biomarkers
FMS-Like Tyrosine Kinase 3 (FLT3)
FMS-like tyrosine kinase 3 is a transmembrane ligand-activated receptor tyrosine, expressed by the hematopoietic stem or progenitor cells, which regulates the differentiation, proliferation, and survival of these cells. Mutations in the FLT3 gene cause dysregulation of the gentle balance between cell proliferation and differentiation.
FLT3 mutations are found in approximately 30% of newly diagnosed AML cases. FLT3 internal tandem duplication (FLT3- ITD), the most frequent mutation (approximately 25% of AML cases) significantly affects the prognosis of AML. Point mutations in the tyrosine kinase domain (FLT3-TKD) have a lower incidence in AML (approximately 7 – 10% of all cases) and their prognostic value is uncertain [9,21,48,]. FLT3-ITD mutations are associated with high White Blood Ccell (WBC) counts, high percentage of peripheral and bone marrow blasts and represent a negative risk factor for overall survival and Event-Free Survival (EFS) [21,47].
FLT3 detection methods include agarose gel electrophoresis following PCR-electrophoresis and fragment analysis using capillary sequencing [53]. Approximately 25% of FLT3-ITD negative relapses are recorded in patients with FLT3-ITD positivity at the time of diagnosis. FLT3-ITD mutation is unstable and nowadays it is not consider being a reliable MRD marker [35].
The prognosis of AML patients with FLT3-ITD mutations is significantly affected by several other factors: the Allelic Ratio (AR) of FLT3-ITD to wildtype FLT3, karyotype, and the presence of nucleophosmin 1 mutations (NPM1) [9,49].
A higher FLT3-ITD allelic ratio (AR>0.5, FLT3-ITDhigh) is associated with higher relapse rates, refractory disease, and shorter OS [16]. Allogeneic hematopoietic stem cell transplant (allo-HSCT) in the first Complete Remission (CR) shows improved Relapse- Free Survival (RFS) and OS in patients with AR>0.5compared to patients receiving standard intensive chemotherapy with or without autologous HSCT. This effect was not seen in patients with low allelic ratio (AR<0.5, FLT3-ITDlow) [55].
Both FLT3-ITD and FLT3-TKD mutations are associated especially with normal cytogenetics (CN-AML) or t(15;17), and rarely with complex karyotype or CBF-leukemia (CBF-MYH11 and RUNX1-RUNX1T1) [42,47]. Patients with t(15;17) acute promyelocytic leukemia harbouring FLT3 mutations are more likely to present with elevated WBC counts and poorer prognosis than those without mutations [45].
The most recent studies propose that patients with FLT3-ITDlow and NPM1 mutations have similarly favorable outcomes as patients with NPM1 mutations without FLT3-ITD.Conversely, patients with FLT3-ITDhigh and NPM1 wild type have a poor prognosis [14]. These are key changes in revisited risk stratification of AML 2017 (Figure 1) [14]. Allogeneic HSCT in CR1 should benefit for patients with intermediate and high risk AML according to the time-dependent analysis [5].
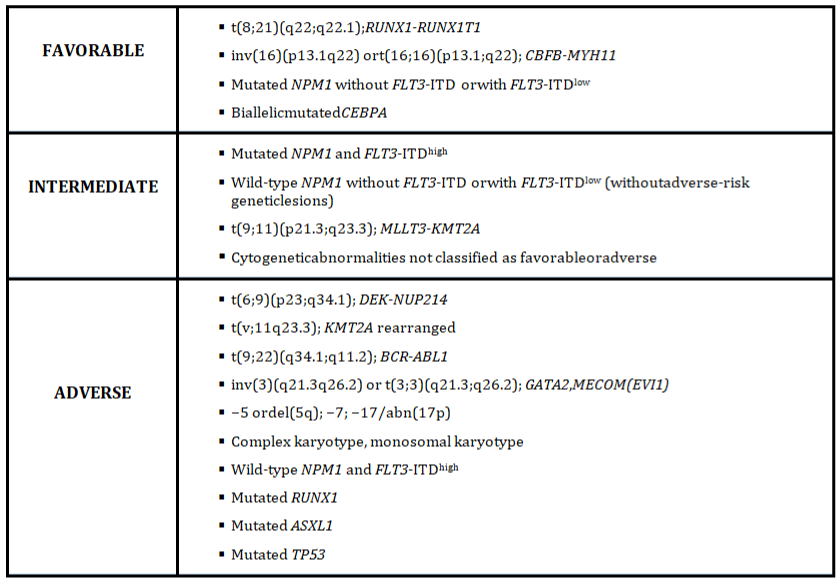
Figure 1: 2017 ELN risk stratification by genetics (Adapted from Döhneret al [14]).
Acknowledgment of FLT3-ITD prognostic impact led to the identification of inhibitors of FLT3 as new therapeutic opportunities in the treatment of AML. Multi-targeted tyrosine kinase inhibitors have shown promising anti leukemic activity in clinical trials [43,62,66]. Midostaurin, the first-generation type I inhibitor, was approved by the FDA in 2017 in combination with standard intensive treatment in FLT3 ITD or TKD mutated AML. The results of the clinical trial have demonstrated a 23% lower risk of death in the midostaurin arm versus placebo [62]. Gilteritinib, the second-generation type I inhibitor, is the other agent approved by the FDA (in 2018) for FLT3-mutated AML. This dual FLT3/ASXL inhibitor is indicated for the relapsed/refractory FLT3 ITD or TKD mutated AML as a single agent. Median OS was 9.3 in the gilteritinib arm versus 5.6 months in the investigator’s choice arm [43].
Nucleophosmin-1 (NPM1)
The nucleophosmin-1 protein regulates multiple cellular events by maintaining genomic stability and DNA repair and apoptosis. The mutation of NPM1 could be critical in malignant transformation [18,48].
NPM1 mutations occur in 30% of AML patients generally in higher median age (46-58 years) [23] and 45-64% of AML patients with NPM1 mutations have a normal karyotype [47]. Mutated NPM1 is associated with good response to standard induction chemotherapy and favorable outcomes without the FLT3-ITD mutation with a high allelic ratio [13,18,47]. The influence of co-occurring mutations with FLT3-ITD is mentioned above. NPM1 mutations are also associated with DNMT3A mutations in almost 60-70% of cases. The presence of DNMT3A mutations most likely negate the favorable effect of NPM1 mutations [18].
Nowadays, RT-qPCR is the most common and sensitive method to determine NPM1. In the future, ddPCR with improved sensitivity allowing for the detection of rare NPM1 mutation subtypes could be used in routine clinical practice [18,28,35,57]. NPM1 mutation is considered to be a reliable MRD marker, these mutations were observed at relapse in nearly all initially NPM1-positive patients. Sensitivity ofNPM1 detection in bone marrow is about 1 log higher than in Peripheral Blood (PB) and time from the first molecular positivity to relapse is longer, if MRD was first detected in bone marrow than in peripheral blood (median, 133 days vs. 87 days) [26]. Balsat et al have mentioned the possibility of diluting the bone marrow sample with peripheral blood and overestimating the quality of their response [3].
Ivey et al have reported that the detection of NPM1 in PBafter the second cycle of intensive chemotherapy was determined to be the optimal time point for MRD [26], while Hubman et al [25] and Balsat et al [3] identified the most appropriate checkpoint to assess the MRD level is in time of recovering hematopoiesis after induction therapy. Patients who achieved <4 log [3] (0.01%) or <3 log (0.1%) [25] Reduction of NPM1 transcripts (used RT-qPCR) in peripheral blood has a higher risk of relapse with inferior survival. MRD positivity seems to be a better predictor of relapse than traditional genetic risk stratification [18]. Therefore, early MRD monitoring of NPM1 mutation in PB may identify AML patients, who will benefit from allergenic HSCT. Duringfollow up, including after allo-HSCT, increasing levels of NPM1 mutation or the change of MRD negativity to MRD positivity nearly always indicated relapse [18]. Interestingly, Balsat et al have observed that NPM1 mutated and FLT3 positive patients, regardless of the allelic ratio, with > 4 log reduction of NPM1 transcript did not benefit from allo-HSCT in CR1 [3].
Isocitratede Hydrogenase 1 and 2 (IDH1/IDH2)
Isocitrate dehydrogenase 1 and 2 are NADP+-depend entenzymes, which catalyze the oxidative decarboxylation of isocitrate to produce CO2 and a-ketoglutarate (aKG). These play a key role in cellular respiration and defense against oxidative stress (Ragon and DiNardo 2017).IDH1/IDH2 mutations induce changes resulting in abnormal enzymatic function and production of an oncometabolite, 2-Hydroxyglutarate (2-HG), which leads to DNA hypermethylation, aberrant gene expression, cell proliferation and abnormal differentiation [38].
IDH mutations (mIDH), specifically IDH1-R132, IDH2-R140, and IDH2-R172, are observed in 15-20% of all AML patients and in 25-30% of patients with a normal cytogenetics [13,47,48]. Patients with IDH mutations are typically older (median age 67). Higher platelet count, a higher percentage of peripheral and bone marrow blasts, and more pronounced neutropenia were detected at the time of diagnosis [38].
Several methods, including PCR, Sanger sequencingor NGS, are commonly used for the detection of IDH mutation [36]. Several publications have described IDH as a potential marker for MRD monitoring [7,10,44,64]. IDH1/IDH2 are markersthat show a response to treatment and reflect the onset of relapse. IDH1/IDH2 can be used as suitable markers for MRD monitoring if there is no other more sensitive marker (e.g. NPM1) [44].
Molecular marker
Prevalence
Tissue
Prognostic value
Utility in MRD
FLT3-ITD
25%
BM
poor
no
FLT3-TKD
7-10%
BM
controversial
no
NPM1
30%
BM/PB1
favorable
yes
IDH1/IDH2
25-30%
BM
controversial
possible
DNMT3A
18-22%
BM
poor
no
CBF leukemias:
RUNX1-RUNX1T1
CBFB-MYH1115-20%
BM
favorable
yes
WT1 expression
80%
PB
poor
Yes
1PB can substitute for BM
Table 1: Molecular markers in AML: prevalence, tissue, prognostic value, utility in MRD.
The prognostic impact of IDH-mutant AML is controversial but seems to be affected by co-mutational status and the specific location of the mutation [38]. Three meta-analyses performed by Feng, Zhou, Xu and colleagues showed that IDH1 mutations were associated with shorter OS and EFS, especially in patients with CN-AML [19,68,71]. Patients with IDH1 single-nucleotide polymorphism (SNP) r11554137 also had shorter OS [68]. Conversely, IDH2 mutations were associated with better prognosis, especially in intermediate-risk AML (frequently trisomy 8) [68]. A study by Papaemmanuilet al [39] has reported a favorable prognosis of the IDH2 R172 mutation, but meta-analysis by Xu et al [68] has shown that prognostic impact has significant heterogeneity and more studies are required to clarify its influence. The prognostic impact of combining with other mutations, especially NPM1, FLT3-ITD, DNMT3A is ambiguous [36,38,68].
IDH1/IDH2 represent prognostic markers for treatment with hypomethylating agents. These mutations are associated with favorable outcomes and significantly higher clinical remission rates during treatment with decitabine or azacitidine [47,48].
It was found that IDH-mutant AML cells are sensitive to the BCL-2 inhibitor venetoclax. Promising data have been reported on treatment with venetoclax combined with Hypomethylating Agents (HMA) in elderly, previously untreated patients with AML. The rate of complete remission and CR with incomplete hematological recovery (CRi) was 67% (97/145) with a tolerable safety profile. Overall survival exceeded 17 months. Patients with IDH1/2 had a median survival rate of 24.4 months, which may show a higher sensitivity of venetoclax in IDH-mutated patients [13,65].
Several selective inhibitors of mutant IDH are now being developed. Enasidenib (AG-221) is the first mutant IDH inhibitor approved by FDA (August 2017) for relapsed/refractory (R/R) IDH2- mutated AML in monotherapy. Enasidenib has shown activity against both IDH2-R140 and IDH2-R172 mutations. Clinical efficacy was observed in 285 patients with IDH2-mutated R/R AML in aphase 1/2 clinical trial. Overall Response Rate (ORR) was 40.3% (114/285), CR was achieved in 19.3% (55/285) with median OS 19.7 months [61]. The first-in-class mutant IDH1 inhibitor ivosidenib (AG-220) was approved in July 2018 as a single agent in IDH1-mutated R/R AML. This approval was based on a large phase 1 dose-escalation trial; in the primary efficacy population CR or CR with partial hematologic recovery was achieved in 30.4% (38/125), and ORR in 41.6% (52/125) [12].
DNA Methyltransferase3A (DNMT3A)
DNA methylation by de novo DNMT3A and 3B (DNMT3B) is an important epigenetic mechanism for genome regulation and development. Dysregulation of DNMT3A and DNMT3B leads to various diseases including hematological malignancies [70]. Mutation at arginine 882 (R882) represents almost 50% of DNMT3A mutations in AML [22,48].
DNMT3A mutations are the most frequent recurrent alterations after FLT3 and NPM1 in AML. DNMT3A mutations occur in 18- 22% of all AML patients. These mutations are highly frequent in the group of patients who have an intermediate-risk cytogenetic profile. Patients with DNMT3A mutated AML compared to DNMT3A wild type AML are older, with higher white blood cell count and are diagnosed with myelomonocytic or monocytic AML [6,29,48].
Sanger sequencing is widely used in the detection of DNMT3A, but its sensitivity is low. Allelespecific PCR orRT-qPCR have a relatively high sensitivity in the detection of these mutations [30]. DNMT3A is not a suitable marker for MRD monitoring. DNMT3A mutations are detectable in patients with long-lasting complete remission, but likely present the persistence of preleukemic clones instead of true leukemic cells. Several studies have presented no clear association between the persistence of DNMT3A mutations in CR and worse prognosis [6].
A large number of studies have presented inferior outcomes of DNMT3A mutated AML compared with DNMT3A wildtype AML [20,29,33,51,59]. Ribeiro et al reported that mutated DNMT3A worsens OS independent of a number of NPM1, FLT3, or CEBPA mutations, and regardless of WBC count, cytogenetic risk, and age [51]. A different study observed that the prognostic impact of DNMT3A mutations depends on age and the type of mutation (R882- DNMT3A or non- R882 DNMT3A). Only non-R882 DNMT3A mutations were associated with poor prognosis in younger patients, while in older patients only R882-DNMT3A mutations had worse outcomes [33].
Despite the above findings, the impact of DNMT3A mutations in clinical decision-making is still unclear, especially for the indication of more aggressive therapeutic strategies such as bone marrow transplantation in the first complete remission of these patients [6]. Patel et al found that intensification of the anthracycline dose (90 mg/ m2/day of daunorubicin) significantly improved outcomes in patients with DNMT3A mutated AML [40]. The same findings were observed in a different group [58]. However, larger prospective clinical trials are necessary for this confirmation [6].
Older patients with DNMT3A mutations have an improved response to treatment by specific DNA methyltransferase inhibitors like azacytidine or decitabine. A higher clinical remission rate and superior OS was achieved by treatment with decitabine in patients with DNMT3A mutations compared to the DNMT3A wildtype mutation (75% vs 34% and 15.2 vs 11 months). However, this comparison is limited by the small number of DNMT3A mutated patients [33].
Core Binding Factor (CBF)
Core binding factor –AML comprises AML with t(8;21) (q22;q22) or inv(16) (p13;q22)/t(16/16) (p13;q22), which generate the RUNX1-RUNX1T1 (AML1-ETO) and CBF-MYH11 fusion genes, respectively [48]. Both alterations disrupt the normal function of the heterodimeric transcription factor CBF complex, which regulates the expression of genes required for normal hematopoiesis [17].
CBF-AML occurs in 15-20% of adult de novo AML cases, predominantly in patients under 60 years of age (patients with inv (16) are generally older than those with t(8;21)). Patients with CBFAML often have a higher marrow blast percentage, hyperleukocytosis, and more frequently, extramedullary disease. Translocation (8;21) is more often associated with AML M2 subtype, while CBF-MYH11 is associated with AML M4Eo. [48] These leukemias are categorized under AML with recurrent genetic abnormalities according to the 2016 WHO classification and are included in the favorable genetic risk group according to the 2017 ELN risk stratification [14].
The transcripts can be detected with RT-qPCR and serve as powerful MRD markers [18,48].
The presence of CBF-AML mutations was found to be associated with a higher remission rate (80-90%) and better OS, and patients benefit from high dose cytarabine post-remission therapy. Nevertheless, approximately 40-45% of these patients relapse [48].
A variety of studies have observed the prognostic significance of CBF-transcripts, but optimal time-points, specimens, and cut-offs are still uncertain. Patients who achieved after one cycle of intensive chemotherapy MRD copy numbers <100 in BM or < 10 in PB for CBFMYH11 AML and <500 in BM or < 100 in PB (or >3 log BM MRD reduction) for RUNX1-RUNX1T1 AML are associated with a lower risk of relapse and better OS (normalized to 105 ABL1 copies). After consolidation therapy and during remission follow up (normalized to 105 ABL1 copies), the MRD detection of mutation gene copy numbers > 50 in BM and > 10 in PB for CBF-MYH11 AML and > 500 in BM and > 100 in PB for RUNX1-RUNX1T1 was associated with relapse in all cases [69]. In another study, the achievement of at least two negative MRD samples in BM for CBF-MYH11 AML during consolidation or early follow-up (≥3 months after complete therapy) was associated with a low risk of relapse (<10%) [18]. Yin et al have reported the median times of molecular to hematological relapse to be 4.9 months for BM and 4.5 months for PB in patients with RUNX1-RUNX1T1 mutation, and 3 months in CBF-MYH11 mutation [69].
In conclusion, MRD detection after induction or completion of consolidation chemotherapy could help to decide about allo-HSCT in patients with insufficient decrease of MRD, but there is still no proof about improved survival with allo-HSCT. It is recommended to measure MRD at least every 3 months in the BM during the first 2 years for both CBF-AML subgroups. Ideally, MRD should be detected in both peripheral blood and bone marrow. But as of yet, PB cannot be considered to be a substitute for BM in clinical practice because of false-negatives [18].
Wilms Tumor Gene 1 (WT1)
The Wilms tumor gene 1, located onchromosome 11p13, is a tumor suppressor gene encoding a zing-finger transcriptional factor that controls cellular growth and differentiation. WT1 expression occurs in CD34+ progenitors and is absent in mature leukocytes in normal adult hematopoiesis [2,31,60]. Mutations in the WT1 gene or its over expression are frequent alterations in myeloid malignancies and can cause deregulation of the WT1 function, which leads to enhanced cell proliferation and hampered cell differentiation [34]. WT1 gene expression indicated poor prognosis in AML patients with NPM1 or FLT3 wild types [15].
WT1 is expressed in more than 80% of adult AML patients in both Bone Marrow (BM) and Peripheral Blood (PB). WT1 expression decreases in patients in remission and increases before hematological relapse. Therefore, WT1 is a suitable marker for monitoring MRD after chemotherapy or HSCT, especially in patients without specific molecular markers. RT-qPCR is the main method of WT1 detection [31,46].
Several studies reported improved OS and EFS when patients had achieved more than a 2log reduction of WT1 expression levels within 61 and 180 days from the start of induction chemotherapy. A standardized WT1 assay after induction therapy could be used to determine AML risk stratification and decision about allo-HSCT in first complete remission [52].
Positive MRD by WT1 expression (MRDWT1) in patients 3 months after ASCT is more often associated with post-transplant relapse, decreased EFS, and poor OS. Correlation between relapse and MRDWT1 is stronger in PB than BM. Positive MRDWT1 is defined as >250 and > 50 copies per 104 ABL copies in BM and PB according to the ELN criteria. Interestingly, 3-month chimerism had less impact on relapse than MRDWT1. Identification of poor risk patients after allo-HSCT by WT1 expression in PB could lead to early immunosuppressive therapy discontinuation or could indicate preemptive donor lymphocyte injection and/or chemotherapy [16].
WT1 mutations have been found in approximately 10% of AML cases with CN-AML. WT1 mutations occur with FLT3 mutation in most cases and lead to induction therapy failure. The level of WT1 after induction therapy helps determine high risk patients for early relapse, those who need more intensive post-induction treatment [47].
Practical Aspects
The ELN MRD working group recommends determining MRD after two cycles of chemotherapy and at the end of treatment. In patients undergoing allo-HSCT, MRD should be measured 4 weeks before allo-HSCT and every 3 months for the first 2 years in BM and PB during the follow-up phase [28].
MRD detection in BM is of higher sensitivity than in PB. However, PB sampling is less burden some for the patient and allows determining MRD level in shorter intervals and therefore detecting earlier the recurrence of disease [28].
MRD by RT-qPCR should be used for patients with positive CBFMYH1, RUNX1-RUNX1T1, and NPM1 mutations. WT1 expression can be detected by RT-qPCR as a potential MRD marker for AML patients without specific mutations. In all other patients, MRD should be detected by MFC [24,57].
MRD threshold highly depends on the applied method, tissue, and analyzed target. MFC-MRD has a recommended cut-off of 0.1%, but lower levels of MRD still have the potential to cause relapse. Regarding RT-qPCR, both absolute thresholds (e.g.,“negativity”, 0.01% or 0.1%) and log reduction to baseline levels at diagnosis time have been determined as clinically relevant. For NGS assays, distinct variant allele frequency level (“negativity”, 0.2% or 2.5 %) have been suggested as MRD thresholds [28,57].
In case of a positive MRD result, confirmation is recommended after 2-4 weeks before the decision of pre-emptive treatment [28].
Conclusion
Genomic alterations are considered to be important markers for risk stratification, MRD monitoring, and treatment-decision making in AML patients. An understandingoftheprognosticsignificanceof these alterations led to theresearchoftargeteddrugs and shiftedthedevelopmentof AML treatment. MRD testing permits therapy modulation or early intervention in patients who do not respond to treatment or relapse early.
In patients with low or intermediate risk at the time of diagnosis, early MRD assessment can help to identify those who could benefit from allo-HSCT. MRD positivity seems to be a better predictor of relapse than pre-therapeutic risk assessment, especially in NPM1 positive AML [3,25,26]. This is similar to patients with acute lymphoblastic leukemia in complete remission after treatment on the GMALL protocol in week 16 with MRD level > 10-4 who should undergo HSCT [25].
This finding raises the need for more prospective randomized clinical trials monitoring MRD, with a subsequent individualized approach to patient treatment including the assessment of the risk of relapse and risk of mortality after allogeneic stem cell transplantation.
Funding
This work has been supported the European Regional Development Fund–Project ENOCH (No. CZ.02.1.01/0.0/0.0/ 16_019/0000868), and the Ministry of Health of the Czech Republic (AZV 17-30089A), Institutional Support by MH CZDRO-FNOs/2019, MH CZ-DROFNOs/ 2020 Student’s grant system SGS15/PrF/2021 University of Ostrava and by The Ministry of Education, Youth and Sports from the Large Infrastructures for Research, Experimental Development and Innovations project e-Infrastructure CZ–LM2018140.
Acknowledgments
The authors would like to give thanks to Shira Timilsina Godfrey, M.D. for editing the article.
Conflict of Interest
The authors declare no conflict of interest.
References
- Alonso Carmen M, Marta Llop, Claudia Sargas, Laia Pedrola, Joaquín Panadero, et al. Clinical Utility of a Next-Generation Sequencing Panel for Acute Myeloid Leukemia Diagnostics. The Journal of Molecular Diagnostics. 2019; 21: 228–40.
- Aref Salah, Solafa El Sharawy, Mohamed Sabry, Emad Azmy, Dalia Abdel Raouf. Prognostic Relevance of Wilms Tumor 1 (WT1) Gene Exon 7 Mutations in-Patient with Cytogenetically Normal Acute Myeloid Leukemia. Indian Journal of Hematology and Blood Transfusion. 2014; 30: 226–30.
- Balsat Marie, Aline Renneville, Xavier Thomas, Stéphane de Botton, Denis Caillot, et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit From Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia With NPM1 Mutation: A Study by the Acute Leukemia French Association Group. Journal of Clinical Oncology. 2017; 35: 185–93.
- Benkova Katerina, Jana Mihalyova, Roman Hajek, Tomas Jelinek. Selinexor, Selective Inhibitor of Nuclear Export: Unselective Bullet for Blood Cancers. Blood Reviews. 2021; 100758.
- Boddu Prajwal C, Tapan M Kadia, Guillermo Garcia-Manero, Jorge Cortes, Mansour Alfayez, et al. Validation of the 2017 European LeukemiaNet Classification for Acute Myeloid Leukemia with NPM1 and FLT3 internal Tandem Duplication Genotypes. Cancer. 2019; 125: 1091–1100.
- Brunetti Lorenzo, Michael C Gundry, Margaret A Goodell. DNMT3A in Leukemia. Cold Spring Harbor Perspectives in Medicine. 2017; 7: a030320.
- Chou W-C, W-C Lei, B-S Ko, H-A Hou, C-Y Chen, et al. The Prognostic Impact and Stability of Isocitrate Dehydrogenase 2 Mutation in Adult Patients with Acute Myeloid Leukemia. Leukemia. 2011; 25: 246–53.
- Cilloni, Daniela, Jessica Petiti, Valentina Rosso, Giacomo Andreani, Matteo Dragani, et al. Digital PCR in Myeloid Malignancies: Ready to Replace Quantitative PCR?. International Journal of Molecular Sciences 2019; 20: 2249.
- Daver Naval, Richard F Schlenk, Nigel H Russell, Mark J Levis. Targeting FLT3 Mutations in AML: Review of Current Knowledge and Evidence. Leukemia. 2019; 33: 299–312.
- Debarri Houria, Delphine Lebon, Christophe Roumier, Meyling Cheok, Alice Marceau-Renaut, et al. IDH1/2 but Not DNMT3A Mutations Are Suitable Targets for Minimal Residual Disease Monitoring in Acute Myeloid Leukemia Patients: A Study by the Acute Leukemia French Association. Oncotarget. 2015; 6: 42345–53.
- Di Nardo Courtney D, Keith Pratz, Vinod Pullarkat, Brian A Jonas, Martha Arellano, et al. Venetoclax Combined with Decitabine or Azacitidine in Treatment-Naive, Elderly Patients with Acute Myeloid Leukemia. Blood. 2019; 133: 7–17.
- DiNardo Courtney D, Eytan M Stein, Stéphane de Botton, Gail J Roboz, Jessica K Altman, et al. Durable Remissions with Ivosidenib in IDH1 -Mutated Relapsed or Refractory AML. New England Journal of Medicine. 2018; 378: 2386–98.
- Di Nardo, Courtney, Curtis Lachowiez. Acute Myeloid Leukemia: From Mutation Profiling to Treatment Decisions. Current Hematologic Malignancy Reports. 2019; 14: 386-394.
- Döhner Hartmut, Elihu Estey, David Grimwade, Sergio Amadori, Frederick R Appelbaum, et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood. 2017; 129: 424–47.
- Du Dongfen, Lixia Zhu, Yungui Wang, Xiujin Ye. Expression of WT1 gene and its prognostic value in patients with acute myeloid leukemia. Zhejiang Da Xue Xue Bao. Yi Xue Ban = Journal of Zhejiang University. Medical Sciences. 2019; 48: 50–57.
- Duléry R, O Nibourel, J Gauthier, V Elsermans, H Behal, et al. Impact of Wilms’ Tumor 1 Expression on Outcome of Patients Undergoing Allogeneic Stem Cell Transplantation for AML. Bone Marrow Transplantation. 2017; 52: 539–43.
- Duployez Nicolas, Alice Marceau-Renaut, Nicolas Boissel, Arnaud Petit, Maxime Bucci, et al. Comprehensive Mutational Profiling of Core Binding Factor Acute Myeloid Leukemia. Blood. 2016; 127: 2451–59.
- Ehinger Mats, Louise Pettersson. Measurable Residual Disease Testing for Personalized Treatment of Acute Myeloid Leukemia. APMIS. 2019; 127: 337–51.
- Feng Jian-Hua, Xiao-Ping Guo, Yuan-Yuan Chen, Zhu-Jun Wang, Yu-Ping Cheng, et al. Prognostic Significance of IDH1 Mutations in Acute Myeloid Leukemia: A Meta-Analysis. American Journal of Blood Research. 2012; 2: 254–64.
- Gale Rosemary E, Katarina Lamb, Christopher Allen, Dima El-Sharkawi, Cassandra Stowe, et al. Simpson’s Paradox and the Impact of Different DNMT3A Mutations on Outcome in Younger Adults With Acute Myeloid Leukemia. Journal of Clinical Oncology. 2015; 33: 2072–83.
- Grafone Tiziana, Michela Palmisano, Chiara Nicci, Sergio Storti. An Overview on the Role of FLT3-Tyrosine Kinase Receptor in Acute Myeloid Leukemia: Biology and Treatment. Oncology Reviews. 2012; 6: 8.
- Guryanova Olga A, Kaitlyn Shank, Barbara Spitzer, Luisa Luciani, Richard P Koche, et al. DNMT3A Mutations Promote Anthracycline Resistance in Acute Myeloid Leukemia via Impaired Nucleosome Remodeling. Nature Medicine. 2016; 22: 1488–95.
- Heath E M, S M Chan, M D Minden, T Murphy, L I Shlush, et al. Biological and Clinical Consequences of NPM1 Mutations in AML. Leukemia. 2017; 31: 798–807.
- Heuser Michael, Alain Mina, Eytan M Stein, Jessica K Altman. How Precision Medicine Is Changing Acute Myeloid Leukemia Therapy.” American Society of Clinical Oncology Educational Book. 2019; 39: 411–20.
- Hubmann M, T Kohnke, E Hoster, S Schneider, A Dufour, et al. Molecular Response Assessment by Quantitative Real-Time Polymerase Chain Reaction after Induction Therapy in NPM1-Mutated Patients Identifies Those at High Risk of Relapse. Haematologica. 2014; 99: 1317–25.
- Ivey Adam, Robert K Hills, Michael A Simpson, Jelena V Jovanovic, Amanda Gilkes, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. New England Journal of Medicine. 2016; 374: 422–33.
- Jelinek T, R Bezdekova, M Zatopkova, L Burgos, M Simicek, et al. Current Applications of Multiparameter Flow Cytometry in Plasma Cell Disorders. Blood Cancer Journal. 2017; 7: e617–e617.
- Jentzsch Madlen, Sebastian Schwind, Enrica Bach, Sebastian Stasik, Christian Thiede, et al. Clinical Challenges and Consequences of Measurable Residual Disease in Non-APL Acute Myeloid Leukemia. Cancers. 2019; 11: 1625.
- Ley Timothy J, Li Ding, Matthew J Walter, Michael D McLellan, Tamara Lamprecht, et al. DNMT3A Mutations in Acute Myeloid Leukemia. New England Journal of Medicine. 2010; 363: 2424–33.
- Li Yunlong, Baosheng Zhu. Acute Myeloid Leukemia with DNMT3A Mutations. Leukemia & Lymphoma. 2014; 55: 2002–12.
- Long Jianting, Shi Fang, Qiangsheng Dai, Xiaolian Liu, Wanshou Zhu, et al. The Wilms Tumor-1 (WT1) Rs16754 Polymorphism Is a Prognostic Factor in Acute Myeloid Leukemia (AML): A Meta-Analysis. Oncotarget. 2016a; 7: 32079–87.
- Long J, Fang S, Dai Q, Liu X, Zhu W, et al. The Wilms Tumor-1 (WT1) Rs16754 Polymorphism Is a Prognostic Factor in Acute Myeloid Leukemia (AML): A Meta-Analysis. Oncotarget. 2016b; 7: 32079–87.
- Marcucci Guido, Klaus H Metzeler, Sebastian Schwind, Heiko Becker, Kati Maharry, et al. Age-Related Prognostic Impact of Different Types of DNMT3A Mutations in Adults With Primary Cytogenetically Normal Acute Myeloid Leukemia. Journal of Clinical Oncology. 2012; 30: 742–50.
- Marjanovic Irena, Teodora Karan-Djurasevic, Milena Ugrin, Marijana Virijevic, Ana Vidovic, et al. Use of Wilms Tumor 1 Gene Expression as a Reliable Marker for Prognosis and Minimal Residual Disease Monitoring in Acute Myeloid Leukemia With Normal Karyotype Patients. Clinical Lymphoma Myeloma and Leukemia. 2017; 17: 312–19.
- Maurillo L, Bassan R, Cascavilla N, Ciceri F. Quality of Response in Acute Myeloid Leukemia: The Role of Minimal Residual Disease. Cancers. 2019; 11: 1417.
- Medeiros B C, A T Fathi, C D DiNardo, D A Pollyea, S M Chan, R Swords. Isocitrate Dehydrogenase Mutations in Myeloid Malignancies. Leukemia. 2017; 31: 272–81.
- Metzeler K H, A Walker, S Geyer, R Garzon, R B Klisovic, C D Bloomfield, et al. DNMT3A Mutations and Response to the Hypomethylating Agent Decitabine in Acute Myeloid Leukemia. Leukemia. 2012; 26: 1106–7.
- Montalban-Bravo, Guillermo, Courtney D DiNardo. The Role of IDH Mutations in Acute Myeloid Leukemia. Future Oncology. 2018; 14: 979–93.
- Papaemmanuil Elli, Moritz Gerstung, Lars Bullinger, Verena I Gaidzik, Peter Paschka, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. The New England Journal of Medicine. 2016; 374: 2209–21.
- Patel Jay P, Mithat Gönen, Maria E Figueroa, Hugo Fernandez, Zhuoxin Sun, et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. New England Journal of Medicine. 2012; 366: 1079–89.
- Paterno Giovangiacinto, Maria Ilaria Del Principe, Adriano Venditti. Detection and Management of Acute Myeloid Leukemia Measurable Residual Disease: Is It Standard of Care?. Current Opinion in Hematology. 2020; 27: 81–87.
- Patnaik, Mrinal M. The Importance of FLT3 Mutational Analysis in Acute Myeloid Leukemia. Leukemia & Lymphoma. 2018; 59: 2273–86.
- Perl Alexander E, Giovanni Martinelli, Jorge E Cortes, Andreas Neubauer, Ellin Berman, et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3 -Mutated AML. New England Journal of Medicine. 2019; 381: 1728–40.
- Petrova Lucie, Filip Vrbacky, Miriam Lanska, Alzbeta Zavrelova, Pavel Zak, et al. IDH1 and IDH2 Mutations in Patients with Acute Myeloid Leukemia: Suitable Targets for Minimal Residual Disease Monitoring?. Clinical Biochemistry. 2018; 61: 34–39.
- Picharski Gledson L, Diancarlos P Andrade, Ana Luiza M R Fabro, Luana Lenzi, Fernanda S Tonin, et al. The Impact of Flt3 Gene Mutations in Acute Promyelocytic Leukemia: A Meta-Analysis. Cancers. 2019; 11: 1311.
- Polák Jaroslav, Hana Hájková, Jacqueline Maalaufová-Soukupová, Jana Marková, Cyril Šálek, et al. Estimation of Molecular Upper Remission Limit for Monitoring Minimal Residual Disease in Peripheral Blood of Acute Myeloid Leukemia Patients by WT1 Expression. Experimental and Therapeutic Medicine. 2012; 3: 129–33.
- Pourrajab, Fatemeh, Mohamad Reza Zare-Khormizi, Azam Sadat Hashemi, Seyedhossein Hekmatimoghaddam. Genetic Characterization and Risk Stratification of Acute Myeloid Leukemia. Cancer Management and Research. 2020; 12: 2231–53.
- Prada-Arismendy Jeanette, Johanna C Arroyave, Sarah Röthlisberger. Molecular Biomarkers in Acute Myeloid Leukemia. Blood Reviews. 2017; 31: 63–76.
- Pratcorona Marta, Salut Brunet, Josep Nomdedéu, Josep Maria Ribera, Mar Tormo, et al Favorable Outcome of Patients with Acute Myeloid Leukemia Harboring a Low-Allelic Burden FLT3-ITD Mutation and Concomitant NPM1 Mutation: Relevance to Post-Remission Therapy. Blood. 2013; 121: 2734–38.
- Ragon Brittany Knick, Courtney D Di Nardo. Targeting IDH1 and IDH2 Mutations in Acute Myeloid Leukemia.” Current Hematologic Malignancy Reports. 2017; 12: 537–46.
- Ribeiro Ana Flávia Tibúrcio, Marta Pratcorona, Claudia Erpelinck- Verschueren, Veronika Rockova, Mathijs Sanders, Saman Abbas, et al. Mutant DNMT3A: A Marker of Poor Prognosis in Acute Myeloid Leukemia. Blood. 2012; 119: 5824–31.
- Rossi Giovanni, Maria Marta Minervini, Angelo Michele Carella, Lorella Melillo, Nicola Cascavilla. Wilms’ Tumor Gene (WT1) Expression and Minimal Residual Disease in Acute Myeloid Leukemia. In Wilms Tumor, edited by Marry M. van den Heuvel-Eibrink. Brisbane (AU): Codon Publications. 2016.
- Sakaguchi Masahiro, Nana Nakajima, Hiroki Yamaguchi, Yuho Najima, Katsuhiro Shono, et al. The Sensitivity of the FLT3-ITD Detection Method Is an Important Consideration When Diagnosing Acute Myeloid Leukemia. Leukemia Research Reports. 2020; 13: 100198.
- Sanz Miguel A, David Grimwade, Martin S Tallman, Bob Lowenberg, Pierre Fenaux, et al. Management of Acute Promyelocytic Leukemia: Recommendations from an Expert Panel on Behalf of the European Leukemia Net. Blood. 2009; 113: 1875–91.
- Schlenk Richard F, Sabine Kayser, Lars Bullinger, Guido Kobbe, Jochen Casper, et al. Differential Impact of Allelic Ratio and Insertion Site in FLT3- ITD–Positive AML with Respect to Allogeneic Transplantation. Blood. 2014; 124: 3441–49.
- Schuurhuis Gerrit J, Michael Heuser, Sylvie Freeman, Marie-Christine Béné, Francesco Buccisano, et al. Minimal/Measurable Residual Disease in AML: A Consensus Document from the European Leukemia Net MRD Working Party. Blood. 2018; 131: 1275–91.
- Schwind Sebastian, Madlen Jentzsch, Enrica Bach, Sebastian Stasik, Christian Thiede, Uwe Platzbecker. Use of Minimal Residual Disease in Acute Myeloid Leukemia Therapy. Current Treatment Options in Oncology. 2020; 21: 8.
- Sehgal Alison R, Phyllis A Gimotty, Jianhua Zhao, Jing-Mei Hsu, Robert Daber, et al. DNMT3A Mutational Status Affects the Results of Dose- Escalated Induction Therapy in Acute Myelogenous Leukemia. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research 2015; 21: 1614–20.
- Shen Yang, Yong-Mei Zhu, Xing Fan, Jing-Yi Shi, Qin-Rong Wang, et al. Gene Mutation Patterns and Their Prognostic Impact in a Cohort of 1185 Patients with Acute Myeloid Leukemia. Blood. 2011; 118: 5593–5603.
- Siehl Jm, E Thiel, R Leben, M Reinwald, W Knauf, Hd Menssen. Quantitative Real-Time RT-PCR Detects Elevated Wilms Tumor Gene (WT1) Expression in Autologous Blood Stem Cell Preparations (PBSCs) from Acute Myeloid Leukemia (AML) Patients Indicating Contamination with Leukemic Blasts. Bone Marrow Transplantation. 2002; 29: 379–81.
- Stein Eytan M, Courtney D DiNardo, Daniel A Pollyea, Amir T Fathi, Gail J Roboz, et al. Enasidenib in Mutant IDH2 Relapsed or Refractory Acute Myeloid Leukemia. Blood. 2017; 130: 722–31.
- Stone Richard M, Sumithra J Mandrekar, Ben L Sanford, Kristina Laumann, Susan Geyer, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. New England Journal of Medicine. 2017; 377: 454–64.
- Sung Pamela J, Selina M Luger. Minimal Residual Disease in Acute Myeloid Leukemia. Current Treatment Options in Oncology. 2017; 18: 1.
- Thol Felicitas, Frederik Damm, Katharina Wagner, Gudrun Göhring, Brigitte Schlegelberger, et al. Prognostic Impact of IDH2 Mutations in Cytogenetically Normal Acute Myeloid Leukemia. Blood. 2010; 116: 614–16.
- Thol Felicitas, Arnold Ganser. Treatment of Relapsed Acute Myeloid Leukemia. Current Treatment Options in Oncology. 2020; 21: 66.
- Voso Maria Teresa, Tiziana Ottone, Serena Lavorgna, Adriano Venditti, Luca Maurillo, et al. MRD in AML: The Role of New Techniques. Frontiers in Oncology. 2019; 9: 655.
- Xu Jie, Jeffrey L Jorgensen, Sa A Wang. How Do We Use Multicolor Flow Cytometry to Detect Minimal Residual Disease in Acute Myeloid Leukemia?. Clinics in Laboratory Medicine. 2017; 37: 787–802.
- Xu Qingyu, Yan Li, Na Lv, Yu Jing, Yihan Xu, et al. Correlation Between Isocitrate Dehydrogenase Gene Aberrations and Prognosis of Patients with Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. Clinical Cancer Research. 2017; 23: 4511–22.
- Yin John A Liu, Michelle A O’Brien, Robert K Hills, Sarah B Daly, Keith Wheatley, et al. Minimal Residual Disease Monitoring by Quantitative RTPCR in Core Binding Factor AML Allows Risk Stratification and Predicts Relapse: Results of the United Kingdom MRC AML-15 Trial. Blood. 2012; 120: 2826–35.
- Zhang Zhi-Min, Rui Lu, Pengcheng Wang, Yang Yu, Dongliang Chen, et al. Structural Basis for DNMT3A-Mediated de Novo DNA Methylation. Nature. 2018; 554: 387–91.
- Zhou Kuang-Guo, Li-Jun Jiang, Zhen Shang, Jue Wang, Liang Huang, et al. Potential Application of IDH1 and IDH2 Mutations as Prognostic Indicators in Non-Promyelocytic Acute Myeloid Leukemia: A Meta-Analysis. Leukemia & Lymphoma. 2012; 53: 2423–29.