
Short Communication
J Hepat Res. 2014;1(3): 1017.
Dried Blood Spots (DBS) for Hepatitis C (HCV) Molecular Diagnostic Testing
Gavin A Cloherty1*, Thomas P Young2, Danijela Lucic1, Corklin Steinhart3, Scott Letendre4
1Abbott Molecular, Des Plaines, IL, USA
2Hospital Israelita Albert Einstein, Brazil
3University of California, San Francisco, San Francisco, CA, USA
4ViiV Healthcare, Research Triangle Park, NC, USA
5University of California, San Diego, San Diego, CA, USA
*Corresponding author: Gavin A Cloherty, Abbott Molecular, Des Plaines, IL, 1300 East Touhy Avenue, Des Plaines, IL 60018, USA
Received: October 31, 2014; Accepted: November 19, 2014; Published: November 21, 2014
Abstract
In resource limited settings, the use of DBS for HCV diagnosis and monitoring has become a necessity for improved patient management. Several studies have evaluated the performance of the RealTime HIV-1 assay using the Whatman 903 device however no data to date has been presented on the RealTime HCV viral load and HCV genotype detection. This study evaluated the use of DBS for HCV viral load testing and viral genotyping. A linear response with a coefficient of determination greaterthan 0.99 was observed for 903 between viral loads of 3.16 log IU/m Land 6.16 log IU/mL (Figure1a/b). A 100% detection rate was achieved at HCV RNA down to 3.16 log IU/mL (Figure 2). HCV RNA was stable in DBS at RT and 2–80C for up to 10 weeks which would allow for convenient storage and shipment of patient samples in resource limited settings. The use of DBS with Abbott RealTime HCV viral load and RealTime HCV Genotype II assays may prove clinically useful for the diagnosis of active HCV infection and the selection of appropriate therapeutic regimens.
Keywords: HCV RNA stability; Dried blood spots, Abbott real time HCV; Abbott Real Time HCV GTII
Introduction
HCV is one of the most common blood borne pathogens in the world and a major source of morbidity globally [1]. The majority of cases of HCV are found in resource-limited settings in Asia, Africa and Latin America. In the US there are an estimated 2.7–3.9 million individuals living with HCV [2] while 49.3–64.0 million individuals are living with HCV in Asia [3]. Current diagnostic tests require phlebotomy followed by refrigeration or freezing until the time of testing. This limits the implementation of HCV diagnosis and initiation of therapy in resource-limited settings where venipuncture and cold chain transportation may not be available. The use of specimen type so then the frozen blood products for HCV diagnostic assays may eliminate this logistical limitation and improve clinical management of patients. In resource-limited settings, the collection of DBS represents an alternative which is minimally invasive, does not require at rained phlebotomist and has proven effective in the diagnosis and monitoring of patients with HIV-1 [4-6]. HCV proteins and nucleic acids from DBS have been successfully quantified using immune assay and polymerase chain reaction (PCR) assays [7-11]. Limited data exist for the quantification of HCV RNA from DBS using modified protocols for commercially available viral load assays. With the advent and introduction of more manageable antiviral therapies for the treatment of HCV, HCV RNA testing will increase dramatically with hopes of identifying and linking patients to care. Overcoming logistical hurdles will be essential if infected populations in high-prevalence regions are to have access to life-saving therapies. In clinical settings where phlebo to my and cold chain storage is a challenge, the use of DBS could be considered.
This study evaluated the detect ability and stability of HCV RNA DBS stored at room temperature and at 2-80C utilizing Whatman 903 collection device. Assays were run using the Abbottm 2000 syste and RealTime HCV RNA (RTVL) and RealTime Genotype II (RTGT) assays (Abbott Molecular Diagnostics Des Plaines, IL, USA). Citrated whole blood was collected from ten HCV seropositive patients by a commercial vendor (Pro Med Dx LLC, Norton, MA). Patient plasma was attained by centrifugation of whole blood at1500 rpm for 20 minutes and tested per manufacturer recommendations with RTVL and RTGTII assays to establish the HCV viral load and genotype. RTVL and RTGT plasma controls were included on each genotype run to demonstrate run validity. Standard amplification and detection protocols were used for all samples processed. HCV RNA detect ability and stability in DBS were evaluated using a five-member HCV panel targeting viral loads from 5.16 – 1.16 log10 IU/mL. These panels were generated by spiking citrated whole blood with HCV seropositive plasma followed by serial dilution in HCV sero-negative citrated whole blood. DBS were stored at (RT) or 2 –80 C in zip lock bags with desiccant until testing. HCV RNA detect ability for 903 was assessed by testing multiple replicates of panels with expected viral loads from 5.16–1.16 log10 IU/mL. Stability of HCV on 903 was evaluated by testing two of these panels with expected viral loads of 5.16 and 3.16 log10 IU/mL at regular intervals up to10 weeks following storage.
Each of the two levels was tested with a minimum of 3 replicates Standard deviation of the three replicates did not exceed 0.25 log IU/ mL. For RTVL testing two whole 50ul spots were excised and placed into1.2mL of Abbott mLysis buffer where as for RTGT a single 75ul spot was excised and placed in 1.2 mL of mLysis buffer. Following a 45 minute incubation at RT, the tube was gently mixed and the supernatant was transferred to a sample tube before being processed using the standard CE marked RTVL or RTGT protocol.
A linear response with a coefficient of determination greater than 0.99 was observed for 903 between viral loads of 3.16 log IU/ mL and 6.16 log IU/mL (Figure1 a/b). A 100% detection rate was achieved at HCV RNA down to 3.16 log IU/mL (Figure 2). Viral quantification correlated well with standard plasma results (based on expected values of dilution panels) with an estimated conversion factor of +2.16 log10 IU/mL. The conversion factor is specific to the sample collection / extraction protocol used (2x50ul spots and 1.0 mL extraction protocol) and was calculated by subtracting the measured viral load of the DBS from the viral load in the original blood sample (ie expected VL – observed DBS VL). Although, the samples within this study are limited to genotype 1 and 3, there was no difference in genotype result between 903 and standard plasma protocol (Table 1). HCV RNA GT was not evaluated beyond the initial time point. HCV RNA in 903 DBS was stable with 100% detection at both levels tested from samples stored at RT or at 2–80C for up to 10 weeks (Figure 3).
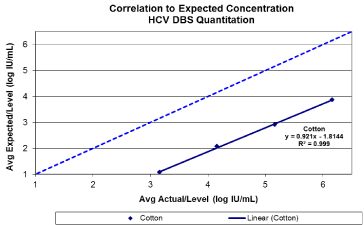
Figure 1(a): Base line HCV viral quantitation from Whatman 903. Target viral
loads between 3-6 log10 IU/ml.
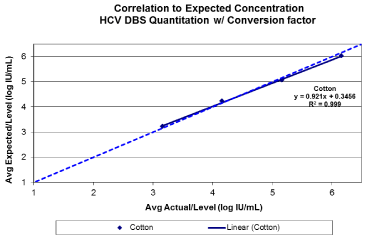
Figure 1(b): Base line HCV viral quantitation from Whatman 903 following
the application of a conversion factor +2.16 log10 IU/mL. Target viral loads
between 3-6 log10 IU/ml.
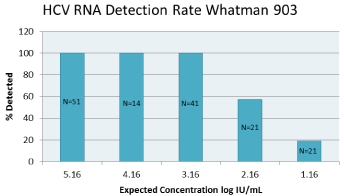
Figure 3: HCVRNA detection rate with Whatman 903. N represents the
number of replicate stested at each level.
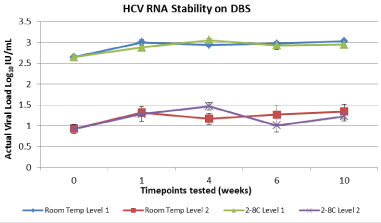
Figure 4: HCV RNA stability onWhatman 903 stored at 2-80C and room
temperature for up to10 weeks.
These findings demonstrate that HCV RNA is stable in DBS at RT and 2–80C for up to10 weeks, which should allow for convenient storage and shipment. Preliminary results indicate that the Abbott RTVL and RTGT assays are compatible with samples collected as DBS. Furthermore, DBS is an effective method for HCV screening and initiation of patients into care in resource-limited settings and in patients with limited venous access. Also, on therapy monitoring of the first generation of DAA’s (Telaprevir) could be implemented by applying the 2.16 log IU/mL conversion factor to the final result. This would allow for monitoring of Telaprevir stopping rules i.e. 1000 IU/mL. Additional optimization of DBS pre-analytical processing steps may increase the sensitivity of the RTVL with this sample type and should be further investigated. This optimization would further allow the DBS device to be used in the SVR assessment of even the second generation DAA’s as most patients that fail therapy or are not adherent to therapy would have a viral load > 3.0 log IU/mL. The use of DBS with Abbott RealTime HCV viral load and RealTime HCV Genotype II assays may prove clinically useful for the diagnosis of active HCV infection, the selection of appropriate therapeutic regimens and confirmation of post treatment sustained virologic response in developing areas with minimal logistic and technical investment.
Acknowledgement
Author contributions: concept and design GAC, TPY, CS, and SL. Data review and analysis, GAC, DL, CS and SL. Manuscript draft, critique, review and approval, GAC, TPY, DL, CS, and SL. Conflict of interest declarations: GAC and DL are employees and shareholders of Abbott Molecular. TPY is a former employee and share holder of Abbott Molecular but has no current conflicts to report. SL has received laboratory materials and support for research protocols. CS has no conflict of interest to report.
References
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005; 5: 558-567.
- Smith BD, Morgan RL, Beckett GA, Falck-Ytter Y, Holtzman D, Teo CG, et al. Recommendations for the identification of chronic hepatitis C virus infection among persons born during 1945-1965. MMWR Recomm Rep. 2012; 61: 1-32.
- Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011; 31 Suppl 2: 61-80.
- Bertagnolio S, Parkin NT, Jordan M, Brooks J, García-Lerma JG. Dried blood spots for HIV-1 drug resistance and viral load testing: A review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev. 2010; 12: 195-208.
- Huang S, Erickson B, Mak WB, Salituro J, Abravaya K. A novel RealTime HIV-1 Qualitative assay for the detection of HIV-1 nucleic acids in dried blood spots and plasma. J Virol Methods. 2011; 178: 216-224.
- Solomon SS, Pulimi S, Rodriguez II, Chaguturu SK, Satish Kumar SK, Mayer KH, et al. Dried blood spots are an acceptable and useful HIV surveillance tool in a remote developing world setting. Int J STD AIDS. 2004; 15: 658-661.
- Bennett S, Gunson RN, McAllister GE, Hutchinson SJ, Goldberg DJ, Cameron SO, et al. Detection of hepatitis C virus RNA in dried blood spots. J Clin Virol. 2012; 54: 106-109.
- Hope VD, Hickman M, Ngui SL, Jones S, Telfer M, Bizzarri M, et al. Measuring the incidence, prevalence and genetic relatedness of hepatitis C infections among a community recruited sample of injecting drug users, using dried blood spots. J Viral Hepat. 2011; 18: 262-270.
- Judd A, Parry J, Hickman M, McDonald T, Jordan L, Lewis K, et al. Evaluation of a modified commercial assay in detecting antibody to hepatitis C virus in oral fluids and dried blood spots. J Med Virol. 2003; 71: 49-55.
- Santos C, Reis A, Dos Santos CV, Damas C, Silva MH, Viana MV, et al. The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. J Virol Methods. 2012; 179: 17-20.
- Solmone M, Girardi E, Costa F, Pucillo L, Ippolito G, Capobianchi MR. Simple and reliable method for detection and genotyping of hepatitis C virus RNA in dried blood spots stored at room temperature. J Clin Microbiol. 2002; 40: 3512-3514.