
Research Article
J Plant Chem and Ecophysiol. 2017; 2(2): 1018.
Characterization of Secondary Metabolites from Endophytic Colletotrichum sp. Isolated from Tragia insuavis
Nyamboki DK1*, Matasyoh JC1 and Wagara IN2
¹Department of Chemistry, Egerton University, Kenya
²Department of Biological Sciences, Egerton University, Kenya
*Corresponding author: Divinah K Nyamboki, Department of Chemistry, Egerton University, P.O. Box 536-20115, Egerton, Kenya
Received: September 25, 2017; Accepted: December 22, 2017; Published: December 29, 2017
Abstract
Antibiotic resistance has persisted over time because of bacterial resistance to available antibiotics. This study sought to isolate and characterize antibacterial secondary metabolites from the crude extracts of Tragia insuavis and two endophytes (TI 2 and TI 3) of the genus Colletotrichum isolated from leaves. The fungal endophytes were screened for bioactivity by carrying out the antagonistic assays against Escherichia coli DSM498 and Staphylococcus aureus ATCC25922. Pure cultures were subjected to solid state fermentation on rice for 21 days; followed by ultra-sonication in methanol and subsequent liquid partitioning of the methanol crude extract between hexane and ethyl acetate. Structural elucidation of the compounds isolated was carried out by a combination of spectroscopic techniques that include 1 and 2D high field NMR spectroscopy and Mass Spectrometry. Antagonistic assays revealed that the fungal endophytes were active against S. aureus (TI 2: 19.67±1.15 mm, TI 3: 22.33±1.53 mm) and E. coli (TI 2: 15.33±1.53 mm, TI 3: 19.67±0.58 mm). 1,2,7-trihydroxyanthracene-9,10-dione (1) and 4-methyl-2-oxopentan-3-yl 2-phenylcyclopropanecarboxylate (2) were isolated from Colletotrichum sp. TI 3 and Colletotrichum sp. TI 2, respectively. The results obtained indicated that Colletotrichum spp. (TI 2 and TI 3) contains compounds that exhibit potential as possible sources of antibacterial agents.
Keywords: Fungal endophytes; Colletotrichum sp; Characterize; Antibiotic resistance
Introduction
Infectious diseases caused by bacteria are one of the major causes of human diseases and deaths in the world. The antibiotic discovery was a relief in the healthcare sector with anticipation that infectious diseases would eventually be minimized [1]. Low and middle income countries have suffered in the public health sector due to antibiotic resistance. Antibiotic resistance is a result of overuse of antibiotics, inappropriate prescriptions and the extensive use of antibiotics in agricultural farming [2]. Measures have been put in place to ensure the effectiveness of antibiotics that are already in the market but implementation has generally been weak. However, antibacterial resistance including multi-drug resistance continues to increase. The development of new antibacterial agents with activity against multidrug resistant bacteria is therefore a critical public health need [3].
Medicines that are obtained from natural sources have played a major role in minimizing and treating human diseases. Different medicinal plant parts are used for extraction of raw drugs as they possess varied medicinal properties [4]. Tragia is a genus of the flowering plants in the spurge family [5]. Various extracts from plants in the genus Tragia have shown pharmacological activity against various ailments that affect human beings [6]. An endophyte is a bacterial or fungal microorganism which spends its life-cycle colonizing inter- and/or intracellularly inside the healthy tissues of the host plant [7]. Endophytic fungi have also been reported as sources of interesting secondary metabolites which include a wide range of compounds such as alkaloids, terpenoids, quinones, peptides, esters, xanthones and phenols [8]. Areas of high biodiversity with a variety of different plant species have a high potential for endophytes with unique secondary metabolites [9].
Secondary metabolites are organic chemical compounds that protect the plant against disease causing pathogens and also help the plant adapt to harsh environmental conditions [10]. These harsh environmental conditions include viruses, fungi, bacteria, mites, nematodes, insects and mammals. These secondary metabolites have a history of protecting humans and animals against disease causing organisms [11]. The ability of plants to produce protective chemical compounds has resulted to the growing interest in herbal medicine [12]. According to [13] and [14], fungal endophytes produce secondary metabolites that protect plants from pathogens and pests. These endophytes are possible sources of lead compounds for the discovery of new drug and therefore should be explored. Some of the genera of fungal endophytes include: Colletotrichum, Fusarium, Aspergillus, Phomopsis, Pestalotiopsis, Neotyphodium and Epichloe among others [15]. Some of the antimicrobial secondary metabolites that have been isolated from Colletotrichum sp. include: 6-isopropenylindole-3-carboxylic acid [16] and colletotric acid [17].
In the present study, endophytic Colletotrichum spp. was isolated from the leaves of T. insuavis. Extraction of secondary metabolites was carried out and the structures of compounds determined.
Multi-drug resistant pathogens have posed challenges in the healthcare sector. Due to these challenges, the need for natural products that can treat these resistant pathogens is compelling. The main object of this study was to isolate bioactive compounds from endophytic Colletotrichum isolated from T. insuavis that can form lead compounds for antibiotic production.
Materials and Methods
Isolation and identification of fungal endophytes
Endophytic fungi were isolated from internal leaf tissues using a method by [18] with slight modification. In this method, the leaves of T. insuavis were washed under running tap water to remove any soil or other foreign materials. The leaves were surface sterilized for 5 minutes using 1% sodium hypochlorite followed by 70% ethanol. Thereafter, the leaves were rinsed twice with sterile distilled water to remove any traces of the disinfectant. The leaves were then cut aseptically into sections approximately 1 mm by 4 mm. The surface sterilized leaves were then plated in petri dishes containing Potato Dextrose Agar (20 g dextrose, 4 g potato extract and 15 g agar) media amended with streptomycin sulphate. The petri dishes were placed in an incubator at 25±2 ºC and monitored for mycelia growth for sub culturing. The isolates were identified by sequencing the Internal Transcribed Spacer (ITS) region of the Ribosomal DNA (rDNA) extracted from the endophytic fungi using automated illumina genome analyzer IIX DNA sequencing machine.
Antagonistic screening of fungal endophytes against pathogenic bacteria
Antagonistic screening of endophytic fungal isolates was done using the dual culture assay following the method described by [19]. The endophytic isolates were grown on PDA medium for 20 days at 25±2 ºC. Plugs of approximately 7 mm were cut using a sterile cork borer and placed in Mueller Hinton agar plates that were seeded with 105 CFU/ml Staphylococcus aureus ATCC25922 (gram positive bacterium) and Escherichia coli DSM498 (gram negative bacterium). The agar plates were incubated at 37 ºC and inhibition zones were measured after 24 hours. The experiment was done in triplicate.
Fermentation of endophytes showing antibacterial activity
Solid fermentation was carried out in ten 500 mL Erlenmeyer flasks containing 90 g of parboiled rice in 90 mL distilled water per flask, previously twice autoclaved at 120 ºC for 40 min for each fungal strain. Agar plugs (about 2 × 2 cm) cut from 7-day-old original cultures on PDA media was used for inoculation. One flask containing autoclaved rice without inoculum was used as control. After 21 days incubation at 30 ºC, 150 mL of methanol was added to each flask and the contents allowed to stand overnight at room temperature. The methanol was filtered and evaporated at reduced pressure, to yield the methanol extract which was submitted to liquid-liquid partitioning between hexane and ethyl acetate. The resulting organic layer was evaporated under reduced pressure to produce hexanic and ethyl acetate extracts [20].
Column chromatography
The dry extract of Colletotrichum sp. TI 3 was re-dissolved in a minimum amount of ethyl acetate and adsorbed on silica gel. The adsorbed sample was loaded on evenly packed silica gel column, carefully to avoid the disturbance of the silica gel layer. Silica gel 60 0.06-0.2 mm (70-230mesh ASTM) supplied by Scharlau Lab supplies Limited was used for the column chromatography. The columns were eluted with ethyl acetate: hexane (5:5) mobile phase. Columns of lengths 50 cm with a diameter of 20 mm were used. Fractions of equal volumes were collected and the TLC of each fraction done. Fractions with similar TLC patterns were grouped together to obtain F1-F5. Preparative High Performance Liquid Chromatography of fraction F4 was done using acidified Milli porewater (H2O + 0.1 HCOOH) and acidified acetonitrile (CH3CN + 0.1 HCOOH). 5.7 mg of compound 2 was obtained.
Purification using Sephadex LH-20
The ethyl acetate extract of Colletotrichum sp. TI 2 was loaded on Sephadex LH-20. HPLC grade methanol was used as the mobile phase. Fractions of equal volumes were collected, and TLC analysis of each fraction done. Fractions of similar TLC patterns were grouped together. The ethyl acetate extract of Colletotrichum sp. TI 2 yielded nine fractions namely: F1-F9. Fraction F8 was further purified using preparative HPLC to obtain compound 1 with a mass of 6.30 mg. Compound 1 was subjected to 1 and 2D high field NMR spectroscopy and mass spectrometry.
Nuclear Magnetic Resonance (NMR) spectroscopy
The 1H, 13C, DEPT, HSQC, COSY and HMBC NMR spectra were recorded on the Bruker Advance 500 MHz NMR spectrometer at the Technical University of Berlin, Germany. The measurements were done in Deuterated DMSO and chemical shifts assigned by comparison with the residue proton and carbon resonance of the solvent. Tetramethylsilane (TMS) was used as an internal standard and chemical shifts were given as δ (ppm). The off- diagonal elements was used to identify the spin - spin coupling interactions in the 1H -1H COSY (Correlation spectroscopy). The proton-carbon connectivity, up to three bonds away, was identified using 1H-13C HMBC (Heteronuclear Multiple Bond Correlation) spectrum. The 1H-13C HSQC spectrum (Heteronuclear Single Quantum Coherence) was used to determine the connectivity of hydrogen to their respective carbon atoms.
Mass spectrometry
The compounds’ mass spectra was recorded on Finnigan Tripple Stage Quadrupol Spectrometer (TSQ-70) with Electron Spray Ionization (ESI) method in the analysis, Thermo Xcalibur Qual computer software was used in analysis of the mass chromatograms.
Data analysis
The mean inhibition zones were calculated and equality of means was analyzed using Statistical Analysis Software (SAS). Turkey’s Honestly Significant Difference (HSD), was used to determine if there was any significant difference between the means of the isolates and the positive control.
Results and Discussion
Antagonistic assay of fungal endophytes against test human pathogenic bacteria
Two endophytes were isolated from the fresh leaves of T. insuavis and identified as Colletotrichum sp. TI 2 and Colletotrichum sp. TI 3. The isolates were screened for their antagonistic activity against E. coli DSM498 and S. aureus ATCC25922 by dual culture method [21] as shown below (Plate 1). Colletotrichum sp. TI 3 was more active against both pathogenic bacteria. Various species of Colletotrichum have been reported to possess bioactivity. Isolates of C. gloeosporioides have been known to show parasitic behavior against fungal pathogens, Pestalotiopsis theae and C. camelliae. The antifungal activity demonstrated by C. gloeosporioides was attributed to the presence of certain diffusible metabolites such as alkaloids, flavonoids, anthraquinones and tannins [22]. According to [23], C. truncatum, an endophyte isolated from Citrus nobilis Lour has been known to inhibit the growth of S. aureus ATCC 25922 and Bacillus subtilis ATCC 6633 but did not inhibit the growth of Pseudomonas aeruginosa ATCC 25932.
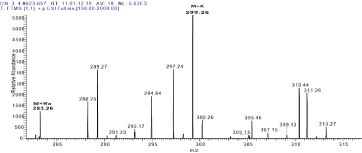
Figure 9: Antagonistic test of endophytic fungi against test pathogenic bacteria.
Plate A shows antagonistic activity of Colletotrichum sp. TI 2 against E. coli, Plate B shows the antagonistic activity of Colletotrichum sp. TI 2 against S. aureus while Plates C and D indicate the antagonistic activity of Colletotrichum sp. TI 3 against E. coli and S. aureus respectively. The endophytes Colletotrichum sp. TI 2 and Colletotrichum sp. TI 3 demonstrated antagonism against both E. coli and S. aureus (Table 1).
Endophytes
Test Organisms (diameter mm, n=3)
Escherichia coli
Staphylococcus aureus
Colletotrichum sp. TI 2
15.33±1.53c
19.67±1.15b
Colletotrichum sp. TI 3
19.67±0.58b
22.33±1.53ab
Chloramphenicol
30.00±0.00a
25.00±0.00a
Table 1: Inhibition diameters (mm) for fungal endophytes against test organisms.
Means with same letter on the same column are not significantly different while those with different letters are significantly different (P<0.05, Turkey’s test).
The inhibition zones of Colletotrichum sp. TI 2 and Colletotrichum sp. TI 3 against E. coli were significantly different in comparison to the reference standard chloramphenicol. Similarly, the inhibition zone of Colletotrichum sp. TI 2 against S. aureus was significantly different from that of chloramphenicol against S. aureus. The inhibition zone of Colletotrichum sp. TI 3 against S. aureus was not significantly different from that of chloramphenicol against S. aureus. This implies that Colletotrichum sp. TI 3 and chloramphenicol have the same level of activity. The endophytes were more active against the gram positive bacteria S. aureus than the gram negative bacteria E. coli. This is because gram negative bacteria have a largely impermeable cell wall and therefore are more resistant to antibiotics. Gram negative bacteria also have efflux pumps that transport antibiotics out of the bacterial cell wall [24]. Colletotrichum sp. TI 3 was more active against both pathogenic bacteria as compared to Colletotrichum sp. TI 2. Therefore, it is anticipated that secondary metabolites from Colletotrichum sp. TI 3 might be having interesting antibacterial compounds.
Structure elucidation of compound 1
Compound 1 (Figure 1) was obtained from Colletotrichum sp. TI 2 as a yellow powder with a mass of 6.29 mg. The DEPT spectrum showed a total of five Methine (CH) carbon atoms resonating at δc - 108.3, 109.1, 127.8, 135.0 and 127.4. The quaternary carbons were resonating at δc - 165.6, 166.0, 109.7, 166.2, 133.4, 185.5, 108.9, 181.7 and 136.0.

Figure 1: Compound 1 with and without HMBC and COSY correlations.
The HSQC spectrum was used to assign protons attached directly to carbon atoms. This spectrum showed correlation between protons resonating at δH- 6.60, 7.13, 8.57, 8.36, 8.26 and C-3, C-4, C-5, C-6 and C-8 respectively.
The HMBC spectrum showed proton correlations with carbon atoms that are two to three bonds away (Figure 1). This gave information on which carbon atoms are next to each other or three bonds away from each other. Proton resonating at δH 6.60 showed correlation with the oxygenated carbon C-1 and the quaternary carbon C-4a which are three bonds away. The proton also showed a two bond correlation with carbon C-4. The proton resonating at δH 7.13 showed correlation with the carbonyl carbon C-10, the oxygenated carbon C-2 and quaternary carbon C-4a, C-9a which are three bonds away and carbon C-3 which is two bonds away. Similarly, protons H-5 (δH 8.57) and H-6 (δH 8.36) showed correlations with carbons C-7, C-10, C-10a, C-6 and carbons C-5, C-7, C-10a respectively. H-8 (δH 8.26) correlated with carbonyl carbon C-9 and quaternary carbons C-8a and C-9a. The carbons C-9 and C-10 were carbonyl carbons characteristic of anthraquinones.
The COSY spectrum identified protons attached to adjacent carbon atoms. The correlation between protons H-3 (δH 6.60) & H-4 (δH 7.13) and H-5 (δH 8.57) & H-6 δH (8.36) showed protons correlated to each other. The 1D and 2D NMR spectral data of compound 1 is shown in Table 2 below. The numbering of compound 1 was done according to [25]. Analytical HPLC indicated that compound 1 had a retention time of 3.9 min as shown in figure 2. The compound absorbs at 880 mAu with a maximum wavelength (λmax) of 215 nm as shown by the corresponding UV spectrum (Figure 3). The High Resolution Electron Impact Mass Spectrometry (HREIMS) of this compound showed ion peaks at m/z 259 that correspond to [C14H8O5 + H]+. The mass spectrum of compound 1 is shown in Figure 4.5.3. Structure elucidation of compound 2
CARBON
13C NMR (DEPT)
1H NMR (HSQC)
TYPE
HMBC
COSY
1
165.6
-
C
-
-
2
166.0
-
C
-
-
3
108.3
6.60
CH
1, 4, 4a
4
4
109.1
7.13
CH
2, 3, 4a, 9a, 10
3
4a
109.7
-
C
-
-
5
127.8
8.57
CH
6, 7, 10, 10a
6
6
135.0
8.36
CH
5, 7, 10a
5
7
166.2
-
C
-
-
8
127.4
8.26
CH
9, 8a, 10a
-
8a
133.4
-
C
-
-
9
185.5
-
C
-
-
9a
108.9
-
C
-
-
10
181.7
-
C
-
-
10a
136.0
-
C
-
-
Table 2: NMR data of compound 1.
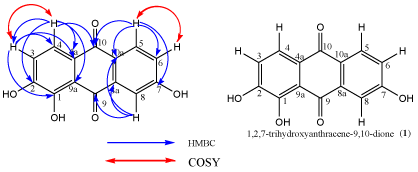
Figure 2: Chromatogram of compound 1.
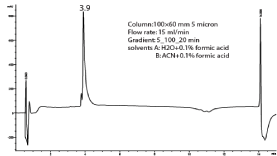
Figure 3: UV spectrum of compound 1.
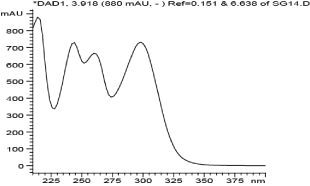
Figure 4: Mass spectrum of compound 1.
Compound 2 (Figure 5) was obtained as a brown powder with a mass of 5.7 mg from Colletotrichum sp. TI 3. The DEPT spectrum, indicated the presence of nine methine carbons (CH) resonating at δC 56.2, 31.8, 75.1, 29.9, 128.8, 129.2, 127.0, 129.2 and 128.8, one methylene carbon (CH2) resonating at δC 34.4 and three methyl carbons (CH3) resonating at δC 31.2, 18.6 and 16.9. The compound contains three quaternary carbons resonating at δC 207.4, 169.1 and 136.2. The 1D and 2D NMR spectral data are summarized in Table 3.
No.
13C (δ)
Type
HSQC (δ)
HMBC
COSY
Coupling constants (J)Hz
1
31.2
CH3
2.09
2
-
S
1'
56.2
CH
5.45
2', 3', 4', 5'
2'
-
2
207.4
C
-
-
-
-
2'
31.8
CH
3.02
1', 4', 5', 6'
1', 3'
-
3
75.1
CH
4.87
4, 4', 6
4
d, J=8.85
3'
34.4
CH2
3.17
1', 5', 6'
2'
dd, J1=4.73, J2=9.76
4
29.9
CH
1.76
3, 6
3, 5, 6
M
4'
169.1
C
-
-
-
-
5
18.6
CH3
0.76
6
4
d, J=6.71
5'
136.2
C
-
-
-
-
6
16.9
CH3
0.24
3, 4, 5
4
d, J=6.72
6'
128.8
CH
7.24
3', 8'
-
-
7'
129.2
CH
7.27
5'
-
-
8'
127.0
CH
7.17
10'
9'
t, J1=6.86, J2=6.87
9'
129.2
CH
7.27
5'
8'
-
10'
128.8
CH
7.24
3', 8'
-
-
Table 3: NMR data of compound 2.
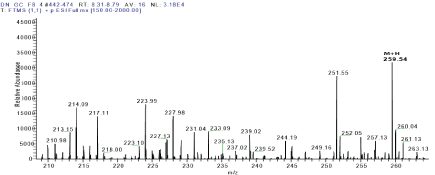
Figure 5: Structure of compound 2 with and without HMBC and COSY
correlations.
The protons attached directly to the carbon atoms were assigned using the HSQC spectrum. This spectrum showed correlation between protons resonating at δH 2.09, 5.45, 3.02, 4.87, 3.17, 1.76, 0.76, 7.24, 7.27, 7.17, 7.27, 7.24 and 0.24 with carbon atoms absorbing at δC 31.2, 56.2, 31.8, 75.1, 34.4, 29.9, 18.6, 128.8, 129.2, 127.0, 129.2, 128.8 and 16.9 respectively. The coupling constants and proton multiplicities were also determined using the proton spectrum. The protons signal at 2.09 ppm was a Singlet (s). Protons H-3 (δ 4.87), H-5 (0.76) and H-11 (0.24) were Doublets (d) with a coupling constant (J) of 8.85 Hz, 6.71 Hz and 6.72 Hz respectively. H-3’ is a Doublet of Doublets (dd) with coupling constants J1 (4.73 Hz) and J2 (9.76 Hz) while H-8’ is a Triplet (t) with coupling constants 6.86 and 6.87 Hz. Proton H-4 is a multiplate. The protons absorbing at δH7.24, 7.27, 7.17, 7.27 and 7.24 were characteristic of protons on an aromatic ring.
The HMBC spectrum showed the correlation between protons and carbons adjacent or three bonds away to each other. H-1 (δ 2.09) correlates with the carbonyl carbon C-2. H-1’ (δ 5.45) correlates with C-2’, C-3’, C-4’ and C-5’. Proton H-2’ (δ 3.02) correlates with C-1’, C-4’, C-5’ and C-6’ while Proton H-3 (δ 4.87) correlates with C-4, C-4’, C-6. The methylene proton H-3’ (δ 3.17) correlates with C-1’ and C-5’. H-4 (δ 1.76) correlates with the methine C-3 and the methyl carbon C-6. The methyl protons H-5 correlates with methyl carbon C-6. The aromatic proton H-6’ (δ 7.24) correlates with C-3’ and C-8’. The proton H-7’ (δ 7.27) correlates with carbon C-5. Proton H-8’ (δ 7.17) correlates with carbon C-10’ which is three bonds away. H-9’ (δ 7.27) correlates with carbon C-5’ which is also three bonds away. The aromatic proton H-10’ (δ 7.24) correlated with C-3’ and C-8’. The methyl proton H-6’ absorbing at δ 0.24 correlates with carbon C-3, C-5 which are three bonds away and C-4 which is two bonds away.
The proton-proton COSY correlations were determined using the COSY spectrum. The correlation between protons H-1’ (δ 5.45) & H-2’ (δ 3.02), H-2’ (δ 3.02) & H-3’ (δ 3.17), H-3 (δ 4.87) & H-4 (δ 1.76), H-4(δ 1.76) & H-5 (δ 0.76), H-4(δ 1.76) & H-6 (δ 0.24) and H-8 (δ 7.17) & H-9 (δ 7.24) were obtained from COSY spectrum. Compound 2 had a retention time of 9.3 min as shown in Figure 6 and its corresponding UV spectrum (Figure 7) indicates that the compound has a maximum wavelength (λmax) at 215 nm with absorption of 848 mAu. The High Resolution Electron Impact Mass Spectrometry (HREIMS) of this compound showed ion peaks at m/z 283 and 299 that correspond to [C16H20O3 + Na]+ and [C16H20O3 + K]+, respectively. The mass spectrum of this compound is shown in Figure 8
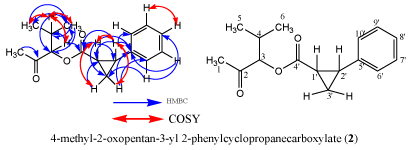
Figure 6: Chromatogram of compound 2.
Bioactivity of secondary metabolites
The bioactivities of compounds isolated in the present study were not determined because they were obtained in small amounts. However, compounds related to 1,2,7-trihydroxyanthracene-9,10- dione isolated from various natural sources have been documented to possess bioactivity. Anthraquinones isolated from Prismatomeris fragrans were reported to possess antimalarial, antifungal and antituberculosis properties at concentrations greater than 20μg/ ml, 50μg/ml and 200μg/ml, respectively [26]. In another study, anthraquinones were isolated from lichen species Xanthoria and exhibited antibacterial activity against Pseudomonas fluorescens, P. glicinea and P. phaseolicola at a concentration of 25μg/disk [27].
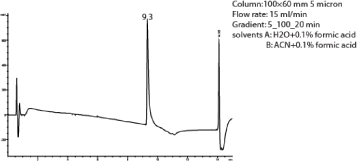
Figure 7: UV spectrum of compound 2.
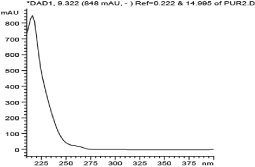
Figure 8: Mass spectrum of compound 2.
Conclusion
In the present study, the cultures of endophytic Colletotrichum spp. (TI 2 and TI 3) showed antibacterial activity against E. coli DSM498 and S. aureus ATCC25922. The class of compounds isolated in this study includes anthraquinones which have been reported to be of medicinal importance. As a result, endophytic fungi Colletotrichum spp. from T. insuavis should be explored as alternative sources of lead compound for antibiotic discovery.
References
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009; 22: 582-610.
- Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015; 40: 277.
- Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL, et al. Critical shortage of new antibiotics in development against multidrug-resistant bacteria-Time to react is now. Drug Resist Updat. 2011; 14: 118-124.
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006; 126: 1037-1048.
- Forster PI. A taxonomic revision of Tragia (Euphorbiaceae) in Australia. Australian Systematic Botany. 1994; 7: 377-383.
- Anthoney ST, Obey JK, Ngule CM. In vitro antibacterial activity of methanolicaqua extract of Plectranthusagentatus leaves. World J Pharmaceutic Res. 2014; 3: 339-349.
- Sturz AV, Christie BR, Nowak J. Bacterial endophytes: potential role in developing sustainable systems of crop production. Critical Reviews in Plant Sciences. 2000; 19: 1-30.
- Yu H, Zhang L, Li L, Zheng C, Guo L, Li W. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol Res. 2010; 165: 437-449.
- Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiology and Molecular Biology Reviews. 2003; 67: 491-502.
- Ngbede J, Yakubu RA, Nyam, DA. Phytochemical screening for active compounds in Canariumschweinfurthii (Atile) leaves from Jos North, Plateau State, Nigeria. Res J Biol Sci. 2008; 3: 1076-1078.
- Ogu GI, Tanimowo WO, Nwachukwu PU, Igere BE. Antimicrobial and phytochemical evaluation of the leaf, stem bark and root extracts of Cyathulaprostrata (L) Blume against some human pathogens. J Intercultural Ethnopharmacology. 2012; 1: 35-43.
- Bandaranayake WM. Quality control, screening, toxicity, and regulation of herbal drugs. Modern phytomedicine: turning medicinal plants into drugs. 2006; 25-57.
- Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, et al. Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences. 2003; 100: 15649-15654.
- Akello J, Dubois T, Gold CS, Coyne D, Nakavuma J, Paparu P. Beauveriabassiana (Balsamo) Vuillemin as an endophyte in tissue culture banana (Musa spp.). J Invertebr Pathol. 2007; 96: 34-42.
- Gimenez C, Cabrera R, Reina M, Gonzalez-Coloma A. Fungal endophytes and their role in plant protection. Current Organic Chemistry. 2007; 11: 707- 720.
- Lu H, Zou WX, Meng JC, Hu J, Tan RX. New bioactive metabolites produced by Colletotrichum sp., an endophytic fungus in Artemisia annua. Plant Science. 2000; 151: 67-73.
- Zou WX, Meng JC, Lu H, Chen GX, Shi GX, Zhang TY. Metabolites of Colletotrichum gloeosporioides, an endophytic fungus in Artemisia mongolica. J Nat Prod. 2000; 63: 1529-1530.
- Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol. 2002; 68: 2198-2208.
- Stadler M, Wollweber H, Fournier J. A host-specific species of Hypoxylon from France, and notes on the chemotaxonomy of The “Hypoxylon rubiginosum complex”. Mycotaxon. 2004; 90: 187-211.
- Nascimento AM, Conti R, Turatti IC, Cavalcanti BC, Costa-Lotufo LV, Pessoa C, et al. Bioactive extracts and chemical constituents of two endophytic strains of Fusarium oxysporum. Revista Brasileira de Farmacognosia. 2012; 22: 1276-1281.
- Campanile G, Ruscelli A, Luisi N. Antagonistic activity of endophytic fungi towards Diplodia corticola assessed by in vitro and in planta tests. European Journal of Plant Pathology. 2007; 117: 237-246.
- Rabha AJ, Naglot A, Sharma GD, Gogoi HK, Veer V. In vitro evaluation of antagonism of endophytic Colletotrichum gloeosporioides against potent fungal pathogens of Camellia sinensis. Indian J Microbiol. 2014; 54: 302-309.
- Hong-Thao PT, Mai-Linh NV, Hong-Lien NT, Van Hieu N. Biological characteristics and antimicrobial activity of endophytic Streptomyces sp. TQR12-4 isolated from Elite Citrus nobilis cultivar ham yen of vietnam. Int J Microbiol. 2016.
- Denyer SP, Maillard JY. Cellular impermeability and uptake of biocides and antibiotics in Gram-negative bacteria. J Appl Microbiol. 2002; 92: 35-45.
- Ruksilp T, Sichaem J, Khumkratok S, Siripong P, Tip-pyang S. Anthraquinones and an iridoid glycoside from the roots of Morindapandurifolia. Biochemical Systematics and Ecology. 2011; 39: 888-892.
- Kanokmedhakul K, Kanokmedhakul S, Phatchana R. Biological activity of Anthraquinones and Triterpenoids from Prismatomerisfragrans. J Ethnopharmacol. 2005; 100: 284-288.
- Manojlovic NT, Solujic S, Sukdolak S, Krstic L. Isolation and antimicrobial activity of anthraquinones from some species of the lichen genus Xanthoria. Journal-Serbian Chemical Society. 2000; 65: 555-560.