
Research Article
Austin J Microbiol. 2016; 2(1): 1010.
Metallo-Beta-Lactamase Producing Gram-Negative Bacteria among Patients Visiting Shahid Gangalal National Heart Centre
Chaudhary AK¹*, Bhandari D², Amatya J¹, Chaudhary P¹ and Acharya B³
¹Department of Microbiology, Tribhuvan University, Nepal
²Public Health Research Laboratory, Institiute of Medicine, Tribhuvan University Teaching Hospital (TUTH), Nepal
³Microbiology Lab, Shahid Gangalal National Heart Centre, Nepal
*Corresponding author: Chaudhary AK, Department of Microbiology, Tri-Chandra Multiple Campus, Tribhuvan University, Ghantaghar, Kathmandu 44600, Nepal
Received: March 01, 2016; Accepted: May 18, 2016; Published: May 20, 2016
Abstract
Background: The rapid spread of acquired Metallo-Beta-Lactamases (MBL) among major Gram-negative pathogens is an emerging threat and a matter of concern worldwide as it results into fewer therapeutic options for the treatment. Therefore, this study was undertaken to determine the prevalence of MBL producing Gram-negative bacteria isolated from different clinical samples.
Methods: A total of 490 samples were analyzed, at the Microbiology Department of Shahid Gangalal National Heart Centre (SGNHC), Bansbari, Kathmandu from December 2013 to June 2014, for routine culture and antibiotic susceptibility testing. MBL detection was done by Imipenem-EDTA Combined Disc Test.
Results: Out of 490 samples analyzed, 107 showed positive growth. Fortytwo percent of the Gram-negative isolates were Multi Drug Resistant (MDR). Among 107 Gram-negative isolates, 66 ceftazidime resistant isolates were screened for MBL production of which 9 (13.6%) were found to be MBL positive. All MBL positive isolates were Pseudomonasaeruginosa. None other Gramnegative bacteria were found to produce MBL. Prevalence of MBL producing P. aeruginosa was 20% and all the isolates were MDR. All the MBL producing P. aeruginosa were isolated from hospitalized patients.
Conclusions: This study showed MBL production in a considerable number of P. aeruginosa isolates with MDR phenotypes. There is a need to track the detection of MBL producers and judicious use of carbapenems is necessary to prevent the further spread of these organisms.
Keywords: MBL; MDR; P. aeruginosa; EDTA; Imipenem
Abbreviations
AST: Antibiotic Susceptibility Test; ATCC: American Type Culture Collection; CLSI: Clinical Laboratory Standard Institute; EDTA: Ethylene-Diamine-Tetraacetic Acid; ESBL: Extended- Spectrum-Beta-Lactamase; ICU: Intensive Care Unit; MBL: Metallo- Beta-Lactamase (Metallo-β-lactamase); MBLs: Metallo-Beta- Lactamases; MDR: Multi-Drug Resistance; SPSS: Statistical Package for Social Science; TUTH: Tribhuvan University Teaching Hospital; ZOI: Zone of Inhibition
Introduction
Metallo-beta-lactamases belong to Amber class B type of Betalactamase and act on a broad spectrum of substrates including penicillins, cephalosporins, and carbapenems [1]. Over the past few years, MBL producing Gram-negative bacteria have emerged as a most widespread and clinically significant carbapenem resistance mechanism [2]. One of the last lines of treatment against high level drug resistant infections is carbapenem, a MBL class of antibiotics, which was developed to resist the beta-lactamase mediated resistance posed by infection causing microbes. However the emergence of New Delhi MBL conferring resistance to almost all beta-lactam antibiotics, including carbapenem, has brought the clinical utility of carbapenem under threat. The production of MBL has recently emerged as one of the most worrisome resistance mechanisms of P. aeruginosa and Acinetobacter species owing to their capacity to hydrolyze all betalactam including Carbapenems [3].
Clinical infections with organisms harboring carbapenemases pose serious therapeutic challenges, with increasing reports of poor patient outcomes and death. So, early detection of resistance strains is crucial which helps in timely implementation of strict infection control practices as well as formulation of clinical guidelines regarding the potential risks for therapeutic failure [4]. This study was conducted with an aim to determine the prevalence of MBL producing Gram-negative bacteriain clinical settings with reference to a hospital in Nepal.
Methods
The present study was conducted at Shahid Gangalal National Heart Centre (SGNHC), Bansbari, Kathmandu from December 2013 to June 2014. Various clinical samples like Urine, Sputum, ET Secretion, Suction Tip, Pus, Wound Swab specimens from both outpatients and in-patients, were included in the study as sent for routine culture to microbiology laboratory and their antibiotic susceptibility testing were performed. Identification of the organisms was carried out following the manual of American Society of Microbiology and Antimicrobial Susceptibility Test (AST) were carried out using Kirby- Bauer disc diffusion method and the result interpreted in compliance with the CLSI (2013) guideline [5].
Identification of the isolates
All clinical isolates were first identified by conventional methods in a routine microbiology laboratory as recommended by the manual of American Society of Microbiology. A positive culture was defined as identification of the organism on Gram-stain followed by growth of the organism in the suitable culture medium.
Identification with staining reactions
Gram-staining was performed for the presumptive identification of the bacteria according to standard technique.
Identification with biochemical test
Typical colonies of bacterial isolates were inoculated on Nutrient broth and incubated at 370C for 4 hours. After incubation, fresh culture of test organism was inoculated into different biochemical media. Test organism was also cultured on Nutrient agar to perform other tests. All the Gram-negative bacteria from clinical isolates were characterized and identified by standard methodology as described in manual of American Society of Microbiology (using a combination of colonial morphology, Gram- stain characteristics, IMViC test, motility test, oxidative-fermentation test, catalase, citrate, oxidase tests and biochemical reactions).
Antimicrobial susceptibility tests
Antibiotic susceptibility test of all isolates was performed by Kirby Bauer disc diffusion method recommended by Clinical Laboratory Standard Institute (CLSI 2013) guidelines using the Mueller-Hinton Agar and recommended antibiotics. In this study, those isolates which acquired non-susceptibility to at least one agent in three or more antimicrobial categories were regarded as MDR [6]. Control strains of E. coli (ATCC 25922) and P. aeruginosa (ATCC 27853) were tested primarily.
Detection of MBL-producers, MBL-screening [7]
The isolates were subjected for MBL detection when the Zone of Inhibition (ZOI) for ceftazidime (30μg) was <18mm. The turbidity of inoculum AST was compared with 0.5 Mc-Farland tube then those screened isolates were subjected for MBL detection.
MBL confirmation test [8]
Two imipenem discs were placed on agar plates containing lawn of test organism. 10 μl of 0.5 M EDTA solution was applied to one of the imipenem disc, placed 25 mm apart (center to center) and the plate was incubated at 370C. After 18-24 hours of incubation, an increase of =7 mm in the zone diameter of imipenem-EDTA disc as compared to imipenem disc alone was considered to be positive test for the presence of MBL.
Data management and analysis
The data from the laboratory finding were entered and analyzed by SPSS version 16.0. Frequency and percentages were calculated and Chi-square test was done whenever applicable with P<0.05 regarded as significant.
Results
Out of the total 490 different clinical samples, 107 (21.83%) samples showed positive growth of Gram-negative bacteria isolates, of which P. aeruginosa 45 (9.18%), E. coli 29 (5.92%), C. diversus 20 (4.08%), K. pneumoniae 10 (2.04%), C. freundi 2 (0.41%) and E. cloacea 1 (0.20%) were isolated.
Gram-negative bacteria from clinical isolates in various culture positive samples
Urine samples showed maximum number of culture positivity of Gram-negative bacteria from clinical isolates, (53.3%), followed by ET Secretion (37.4%), Pus (2.8%), Wound Swab (2.8%), Sputum (1.9%) and Suction Tip (1.9%) (Table 1).
E. coli
P. aeruginosa
C. diversus
C. freundi
E. cloacea
K. pneumoniae
Total
Urine
23
11
19
0
1
3
57
21.5%
10.3%
17.8%
0%
0.9%
2.8%
53.3%
Sputum
0
2
0
0
0
0
2
0%
1.9%
0%
0%
0%
0%
1.9%
ET Secretion
5
28
1
0
0
6
40
4.7%
26.2%
0.9%
.0%
.0%
5.6%
37.4%
Suction Tip
1
0
0
1
0
0
2
0.9%
0%
0%
.9%
.0%
.0%
1.9%
Pus
0
2
0
1
0
0
3
0%
1.9%
0%
0.9%
.0%
.0%
2.8%
Wound Swab
0
2
0
0
0
1
3
0%
1.9%
0%
0%
0%
0.9%
2.8%
Total
29
45
20
2
1
10
107
27.1%
42.1%
18.7%
1.9%
.9%
9.3%
100.0%
Table 1: Gram-negative bacteria from clinical isolates in various culture positive samples (n=107).
Antibiotic susceptibility pattern of Gram-negative bacteria from clinical isolates
The antibiotic susceptibility pattern of Gram-negative bacteria from clinical isolates revealed that the most sensitive drug was nitrofurantoin (89.1%) among Urine samples. The most sensitive drug was piperacillin/tazobactam (82.5%), followed by imipenem (82.1%) and amikacin (71.9%) among samples other than Urine. Similarly, the most resistant drug was ampicillin (87.1%) followed by cephalexin (83.9%), ceftazidime (68.75%) and cefixime (67.7%) respectively.
Antibiotic susceptibility pattern of P. aeruginosa
The antibiotic susceptibility pattern of P. aeruginosa revealed that the maximum sensitivity was observed for piperacillin/tazobactam (86.7%) followed by imipenem (77.8%), amikacin (64.4%), meropenem (60.0%) and piperacillin (60.0%) (Table 2).
Sensitive
Intermediate
Resistant
Amikacin
29
0
16
64.4%
0%
35.6%
Ciprofloxacin
5
3
37
11.1%
6.7%
82.2%
Ceftazidime
3
0
42
6.7%
0%
93.3%
Gentamicin
9
0
36
20.0%
0%
80.0%
Imipenem
35
0
10
77.8%
0%
22.2%
Meropenem
27
1
17
60.0%
2.2%
37.8%
Piperacillin
27
1
17
60.0%
2.2%
37.8%
Piperacillin/Tazobactam
39
0
6
86.7%
0%
13.3%
Table 2: Antibiotic susceptibility pattern of P. aeruginosa (n=45).
Distribution of MDR isolates and MBL production among Gram-negative bacteria from clinical isolates
In this study, out of 107 Gram-negative bacteria from clinical isolates, 45 were found to be MDR and maximum MDR was observed in P. aeruginosa (42.1%), followed by E. coli (27.1%), C. diversus (18.7%), K. pneumoniae (9.3%), C. freundi (1.9%), E. cloacea (0.9%) respectively.
Out of 107 Gram-negative isolates, 9 isolates (8.4%) were MBL producers. The production of MBL was seen only in 20 % P. aeruginosa among 45 isolates (Table 3).
MDR isolates
MBL Production
Positive
Non-MDR
Total
Positive
Negative
Total
E. coli
10
19
29
0
29
29
34.5%
65.5%
27.1%
0%
100.0%
27.1%
P. aeruginosa
26
19
45
9
36
45
57.8%
42.2%
42.1%
20.0%
80.0%
42.1%
C. diversus
1
19
20
0
20
20
5.0%
95.0%
18.7%
0%
100.0%
18.7%
C. freundi
2
0
2
0
2
2
100.0%
0%
1.9%
0%
100.0%
1.9%
E. cloacea
1
0
1
0
1
1
100.0%
0%
0.9%
0%
100.0%
0.9%
K. pneumoniae
5
5
10
0
10
10
50.0%
50.0%
9.3%
0%
100.0%
9.3%
Total
45
62
107
9
98
107
42.1%
57.9%
100.0%
8.4%
91.6%
100.0%
Table 3: MDR isolates and MBL production among Gram-negative bacteria from clinical isolates.
Distribution of MBL positive P. aeruginosa in different clinical samples
Out of 9 MBL positive isolates, higher incidence of MBL producing strains were found among samples of Endo-Tracheal- Secretion (n=6) (Table 4).
Positive
Negative
Total
Urine
2
55
57
22.2%
56.1%
53.3%
Sputum
0
2
2
0%
2.0%
1.9%
ET Secretion
6
34
40
66.7%
34.7%
37.4%
Suction tip
0
2
2
0%
2.0%
1.9%
Pus
0
3
3
0%
3.1%
2.8%
Wound swab
1
2
3
11.1%
2.0%
2.8%
Total
9
98
107
8.4%
91.6%
100.0%
Table 4: MBL positive P. aeruginosa in different clinical samples.
Distribution of MBL producing P. aeruginosa among Inpatient and Out-patient
Isolates from In-patients were found to carry MBL-encoded resistance property. All MBL positive P. aeruginosa were isolated from in-patients only (Figure 1).
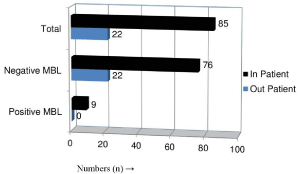
Figure 1: MBL producing P. aeruginosa among In-patient (n=85) and Outpatient
(n=22).
Antibiotic susceptibility pattern of MBL Positive P. aeruginosa
The antibiotic susceptibility pattern of MBL Positive P. aeruginosa revealed that the maximum sensitivity was observed for piperacillin/ tazobactam 77.77% followed by amikacin 44.44%, piperacillin 33.33% and Imipenam 11.11%. Hundred percent resistances were observed for ceftazidime, gentamicin, ciprofloxacin and meropenem (Table 5).
Sensitive
Intermediate
Resistant
Amikacin
4
0
5
44.4%
0%
55.5%
Ciprofloxacin
0
0
9
0%
0%
100.0%
Ceftazidime
0
0
9
0%
0%
100.0%
Gentamicin
0
0
9
0%
0%
100.0%
Imipenem
1
0
8
11.11%
0%
88.88%
Meropenem
0
0
9
0%
0%
100.0%
Piperacillin
3
0
6
33.33%
0%
66.66%
Piperacillin/Tazobactam
7
0
2
77.77%
0%
22.22%
Table 5: Antibiotic susceptibility pattern of MBL Positive P. aeruginosa (n=9).
Discussion
The rapid spread of acquired MBLs among major gram-negative pathogens is a matter of particular concern worldwide and in Nepal as well. This study showed a high incidence of MBL- producing gramnegative bacteria in different clinical samples. In Nepal first study of MBL was conducted at Department of Microbiology, Tribhuvan. University Teaching Hospital (TUTH) from June to November 2008, in which 6 (1.3%), 3 isolates each of P. aeruginosa and Acinetobacter spp. of the total 448 Gram-negative isolates were found to be MBL producer [7]. However, several studies from Nepal and India indicated higher prevalence of MBL producing gram-negative isolates [9-12], which might have resulted from overuse of carbapenems in the hospital settings.
This study showed the increasing rate of antimicrobial resistance among Gram- negative bacteria especially towards ampicillin and Cephalosporins. Among MDR isolates, the most sensitive drug was Piperacillin/Tazobactam, followed by Imipenem and Amikacin. All of MBL producers in this study were MDR and resistant to most of the antibiotic used. This is due to the fact that the MBL genes are often carried along with other resistance genes resulting in multidrug resistance limiting treatment options [13]. Similar results were obtained in previous studies [7,14].
Although, PCR method is simple to use in detecting MBL producing isolates, it has become more difficult with the increased number and types of MBL. Combined disc test is simple to perform and highly sensitive in differentiating MBL-producing isolates. Thus, implementation of simple method using imipenem-EDTA disk for MBL detection is quick, specific, sensitive and reproducible [15]. Out of 107 Gram- negative isolates, 9 isolates (8.4%) were found to be MBL producers. The production of MBL was seen in 20 percent P. aeruginosa among 45 isolates.
In this study, among the 9 MBL-producers, only one isolate was sensitive to imipenem. As seen with extended-spectrum-betalactamase and AmpC beta-lactamases with cephalosporins, MBL carrying organisms may also appear susceptible to carbapenems using current CLSI breakpoints but they may not be effective in in vivo [5]. The method, utilizing ceftazidime resistance as the selective criterion for the phenotype test to detect MBL, was fruitful. If only carbapenems resistant cases were selected, one MBL carrying isolate would have been missed.
Hundred percent resistances to ceftazidime, gentamicin, ciprofloxacin and meropenem were observed among MBL producers, which was consistent with Mishra et al. [7]. In this study 77.77% of MBL producers were susceptible to piperacillin/tazobactam which was similar to the finding of Varaiya et al. [10] and 44.44% were sensitive to amikacin which was nearly similar to the study conducted by Gupta et al. [16].
Emergence of MBL producing P. aeruginosa in intensive care units is alarming and reflects overuse of carbapenems. There is intense selection pressure, due to high usage of broad spectrum antibiotics in ICUs. This results in eradication of competitive flora and subsequent selection of multidrug-resistant strains [11]. All MBL producing P. aeruginosa were isolated from In-patients and out of 9 MBL positive isolates 5 were from Endo Tracheal secretion. De et al. [17] reported prolong hospitalization > 8 days; mechanical ventilation and endotracheal intubation were common risk factors of MBL producing in ICU patients. Zavascki et al. [18] have shown that prior exposure of a Beta-lactam or fluoroquinolone, neurological disease, Urinary Tract Infection, renal failure and ICU stay were significant risk factors for MBL producing infections. This may be the reason behind MBL detection only among Inpatients in this study.
Carbapenems being the effective therapeutic agents against highly resistant pathogens such as Pseudomonas spp. and gram negative bacteria, spread of this resistance among these pathogens and transfer to other Gram-negative bacteria would seriously restrict therapeutic options. The occurrence of an MBL-positive isolate in a hospital setting poses a therapeutic problem because the only therapeutic alternative remains the potentially toxic polymyxinB and colistin. The accurate identification and reporting of MBL-producing bacteria will aid infection control practitioners in preventing the spread of these multidrug-resistant isolates.
Conclusion
Among different Gram-negative bacteria isolated, only P. aeruginosa showed MBL production. None of the other Gramnegative bacteria was found to be MBL positive. This study demonstrates Gram-negative bacteria exhibiting the increased rate of resistance to wide variety of antibiotics from different antimicrobial categories with additional burden of MBL producers within them. All MBL producers are multi-drug-resistant, leaving very few antibiotics for treatment options. This study highlights the need to do regular surveillance for the detection of MBL producers, judiciously use carbapenems to prevent their spread and use effective antibiotics after sensitivity testing for treatment.
Acknowledgements
We would like to thank all the patients whose samples were used in this study, all the staff of Shahid Gangalal National Heart Centre, Bansbari, Kathmandu, all the faculties, and staffs of Department of Microbiology, Tri-Chandra Multiple Campus, Kathmandu, and all our friends.
References
- Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005; 43: 3129-3135.
- Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, et al. New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis. 2013; 19: 870-878.
- Deshmukh DG, Damle AS, Bajaj JK, Bhakre JB, Patwardhan NS. Metallo-β-lactamase-producing clinical isolates from patients of a tertiary care hospital. J Lab Physicians. 2011; 3: 93-97.
- Franklin C, Liolios L, Peleg AY. Phenotypic detection of carbapenem-susceptible metallo-beta-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006; 44: 3139-3144.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA: CLSI; 2007, M100-S17.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18: 268-281.
- Mishra SK, Acharya J, Kattel HP, Koirala J, Rijal BP, Pokhrel BM. Metallo-beta-lactamase producing gram-negative bacterial isolates. J Nepal Health Res Counc. 2012; 10: 208-213.
- Yong D, Lee K, Yum JH, Shin HB, Rossolini GM, Chong Y. Imipenem-EDTA disk method for differentiation of metallo-beta-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2002; 40: 3798-3801.
- Khanal S, Joshi DR, Bhatta DR, Devkota U, Pokhrel BM. β-Lactamase-Producing Multidrug-Resistant Bacterial Pathogens from Tracheal Aspirates of Intensive Care Unit Patients at National Institute of Neurological and Allied Sciences, Nepal. ISRN Microbiol. 2013; 2013: 847569.
- Varaiya A, Kulkarni N, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo beta lactamase producing Pseudomonas aeruginosa in ICU patients. Indian J Med Res. 2008; 127: 398-402.
- Kali A, Srirangaraj S, Kumar S, Divya HA, Kalyani A, Umadevi S. Detection of metallo-beta-lactamase producing Pseudomonas aeruginosa in intensive care units. Australas Med J. 2013; 6: 686-693.
- Kumar SH, De AS, Baveja SM, Gore MA . Prevalence and risk factors of Metallo β-lactamase producing Pseudomonas aeruginosa and Acinetobacter species in burns and surgical wards in a tertiary care hospital. J Lab Physicians. 2012; 4: 39-42.
- Walsh TR. The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria. Clin Microbiol Infect. 2005; 11: 2-9.
- Anuradha K, WadekarMita D, Venkatesha D. Phenotypic detection of ESBL and MBL in clinical isolates of Enterobacteriaceae. 2013; 1: 89-95
- Galani I, Rekatsina PD, Hatzaki D, Plachouras D, Souli M, Giamarellou H. Evaluation of different laboratory tests for the detection of metallo-beta-lactamase production in Enterobacteriaceae. J Antimicrob Chemother. 2008; 61: 548-553.
- Gupta V, Sindhu S, Chander J. MBL producing non-fermentative Gram-negative bacteria: An increasing clinical threat among hospitalized patients. Asian Pac J Trop Med. 2012; 5: 718-721.
- De AS, Kumar SH, Baveja SM . Prevalence of metallo-β-lactamase producing Pseudomonas aeruginosa and Acinetobacter species in intensive care areas in a tertiary care hospital. Indian J Crit Care Med. 2010; 14: 217-219.
- Zavascki AP, Barth AL, Gaspareto PB, Gonçalves AL, Moro AL, Fernandes JF, et al. Risk factors for nosocomial infections due to Pseudomonas aeruginosa producing metallo-beta-lactamase in two tertiary-care teaching hospitals. J Antimicrob Chemother. 2006; 58: 882-885.