
Research Article
Austin J Microbiol. 2019; 5(1): 1024.
Bioremediation of Remazol Blue RGB by Newly Isolated Bacillus Strain
Sweta PB* and Tank SK
Department of Biosciences, Veer Narmad South Gujarat University, Gujarat, India
*Corresponding author: Sweta Parmita Bera, Department of Biosciences, Veer Narmad South Gujarat University, Udhana - Magdalla Road, Surat, Gujarat, India
Received: December 31, 2018; Accepted: February 18, 2019; Published: February 25, 2019
Abstract
Disposal of dyes into the environment causes serious damage and also they may be toxic to some aquatic organisms due to their breakdown products. In the present study, an attempt was made to examine the potential of isolated bacterium for decolorization of Remazol Blue RGB dye in batch reactors. A potential bacterial strain was isolated and selected from the textile effluent on the basis of rapid azo dye Remazol Blue RGB (150 mgl-1) decolorization and later identified as belonging to genus Bacillus based on phenotypic characterization. Isolate 1, 2 and 3 having the competition to degrade Remazol Blue RGB effectively was examined by means of modified mineral salt. Isolate 1 was absolutely competent to remove the dye from the liquid medium at 10h. The isolate 1 showed the most excellent performance at 150 mgL-1 dye concentration (pH 7.5) at a temperature of 37°C. Likewise, yeast extract is the best source of carbon for discoloration purposes. The results imply that the isolate 1 could be used for the elimination of the reactive dyes of the textile effluents. The effect of pH, temperature, carbon & nitrogen source was studied with an aim to determine the optimal conditions required for maximum decolorization and degradation. The results show that the isolated bacterium has a dynamic potential in removal of dye Remazol Blue RGB from wastewater under aerobic conditions.
Keywords: Remazol Blue RGB; Textile effluents; Yeast extract; Bacillus
Introduction
Textile industry effluent is known to contain strong colour, a highly fluctuating pH, significant Chemical oxygen demand, Biological oxygen demand and Total dissolved solids. Effluents from the textile industries are highly coloured containing dyes that vary from 2% for basic dyes to as high as 50% for reactive dyes, leading to severe contamination of surface and ground waters in the vicinity of dyeing industries. Dyes are an important class of synthetic organic compounds, widely used in textile, leather, plastic, cosmetic and food industries and are therefore common industrial pollutants [1]. During dyeing process a large proportion of the applied dyes are discharged as waste effluent to the environment without proper disposal of toxic matters [2]. The textile dyes represent a category of organic compounds, generally considered as pollutants, presented into wastewaters resulting mainly from processes of chemical textile finishing. Dyes also obstruct light penetration and oxygen transfer that affects water bodies [3]. Because of their synthetic nature and structure mainly aromatic, the most of dyes are non-biodegradable, having carcinogenic action or causing allergies, dermatitis, skin irritation or different tissular changes. The textile organic dyes must be separated and eliminated from industrial wastewaters by effective and viable treatment includes different separation processes (sedimentation, filtration, membrane separation), and some physicochemical treatment (i.e. adsorption; coagulation-flocculation with inorganic coagulants and organic polymers; chemical oxidation; ozonation; electrochemical process, etc.). Synthetic dyes are broadly dividing into azo, reactive, triphenylmethane, heterocyclic, polymeric structure etc [4]. From these, Azo dyes are the main constituents of such pollution because of their wide applicability and usages, and therefore, these are present majorly in textile industrial effluents. The characteristics natures of azo compounds are nitrogen to nitrogen double bond (R-N=N-R) and aromatic rings mostly substituted by sulfonate groups [5-7]. The treatment of textile industrial effluent is difficult for the reason of the double bond of azo dyes, which are resisting to breakdown and are toxic, carcinogenic or mutagenic and serious hazard to living organisms. Because of its resistance to degradation by light, chemicals and microbes, textile industry wastewater treatment is very challenging [8-12]. Moreover, their toxicity and resistance to degradation offer great challenge for removal technologies. In many cases, the products formed after the degradation of the parent azo dye molecule are more toxic. A number of biotechnological approaches have attracted interest with regard to tackling textile industrial dye pollution in an eco-efficient manner, mainly with the utilization of bacteria and frequently in amalgamation with physicochemical processes [13-16]. Bioremediation is defined as the process by using microorganism to degrade hazardous pollutants transform into environmental friendly less harmful forms. The use of microbial techniques to deal with pollution or bioremediation is a key research area in the environmental sciences [17,18]. Biological treatment offers a cheaper and environment friendly alternative to dye decolorization and wastewater reutilization in industrial process [19-22]. The general approach for bioremediation of textile effluent is to improve the natural degradation capacity of the indigenous microorganism that allows degradation and mineralization of dyes with a low environmental impact and without using potentially toxic chemical substances, under mild pH and temperature conditions [23- 28].
Materials and Methods
Waste water sample collection and analysis
Surat is known as the ‘Textile capital’of India. The textile industry is one of the oldest and the most widespread industries in Surat. It is ideal for textile waste water sample collection. The samples were collected from the activated sludge of the common textile effluent treatment plant of Surat, Gujarat, India. Samples were collected from the aeration tank of effluent treatment plant. The Temperature and pH was analyzed at the site. The examination of temperature was done using laboratory grade thermometer and pH was analyzed by using pH meter (Hanna digital pH meter). The sample was transferred to laboratory at 4°C as per the standard methods. The physicochemical parameters such as Color, Biological Oxidation Demand, Chemical Oxygen Demand, Total Suspended Solids, and Total Dissolved Solids were analyzed as soon as the sample was brought to the laboratory (Table 1). Sample colour was analyzed by spectrophotometer (SHIMADZU UV-1 800). BOD was determined by employing evaporation method by dissolve oxygen meter while chemical oxygen demand was measured by instrument directly.
Sr. No
Parameter
Unit
Effluent
1
Colour
-
Greyish blue
2
Smell
-
Pungent
3
pH
-
9.2
4
COD
ppm
10800
5
BOD
ppm
3500
6
TSS
ppm
1500
7
TDS
ppm
25700
8
NH4+N
ppm
550
Table 1:
Enrichment and isolation of dye decolorizing bacteria
The bacterial strains were isolated from textile waste water containing activated sludge. A modified MSM medium with Remazol Blue RGB dye as the source of C and N was used to enrich the Inoculums developed from the isolate. Remazol Black B dye was added to a concentration of 150 mg L-1. The cultures containing 200 ml MSM with a dye broth in 500 ml Erlenmeyer flasks were inoculated with 10 ml volume of activated sludge. The flasks were incubated at 37°C for 5 days under static conditions. After incubation, cell suspensions from each vial were loaded onto MSM agar medium and incubated at 37°C for 24 hours. Bacterial colonies that appeared on the agar medium were picked, gently washed with sterile water and resuspended in flasks containing fresh MSM broth enriched with dye. Approximately 14 active colonies were selected for purification.
Purification of bacterial isolates
The selected isolates were purified by streaking on agar containing MSM medium at a concentration of 20 g L-1. Streaking was performed three times in a zigzag manner. The purified cultures were stored in a refrigerator for further study.
Screening
Using a modified MSM medium, primary screening is performed to obtain the perfect bacterial strains capable of discoloring azo dye. For this purpose, 14 isolates capable of bleaching Remazol Blue RGB from all samples were selected. The bleaching ability of each isolate was tested in liquid medium. Modified Remazol Blue RGB medium was incubated at 37°C for 24 hours with the appropriate bacterial inoculum. After 24 hours, cell harvesting was performed by centrifugation at 10,000 rpm (REMI R-23, India) for 10 minutes. Thereafter, decolorization was measured using a spectrophotometer (SHIMADZU-1 800, Japan) at 597 nm. The uninoculated media was processed as a blanks for the control of abiotic decolorization. The three most effective bacterial isolates (Isolate 1, 2 and 3) from the final screening were retested for their bleaching potentials in tubes at different time intervals. 0.5% of the yeast extract as a common substrate was added in 10 ml sterilized MSM broth containing Remozol Blue RGB at a concentration of 150 mg L-1. The uniform cell density of the inoculum was added to the appropriate bacterial strains at (OD: 0.6) 597 nm. The azo-dye containing MSM medium with yeast extract in uninoculated tubes was incubated in parallel media to ensure abiotic discoloration of the dye. The staining was measured every 6 hours of the interval for up to 24 hours at a 597 nm spectrophotometer as described by Khalid et al.
Environmental parameters optimization
Significant parameters such as substrate concentration, temperature and pH were optimized for assays for different carbon sources (Mannitol, Yeast Extract, Glucose, Maltose) at 5 g of L-1 concentration were investigated as a co-substrate in the development of dye removal. Optimization studies included a selection of dye concentrations (50, 75, 100, 125, 150, 200 and 250 mg L-1), temperatures (25, 30, 35, 40, 45°C) [6,7,8,9]. Isolated strains 1, 2 and 3 were designed to optimize their color removal capabilities. Since the culture conditions were unchanged as used in the attempt to remove the colors, i.e., 100 mg L-1 Remazol Blue RGB azo dye with MSM was used. Uninfected blanks were made to ensure abiotic decolorization during experiments.
Identification of selected isolate by 16s rRNA gene sequencing method
For sequencing the 16S rRNA gene, the DNA was isolated using a bacterial DNA isolation kit (Fungal / Bacterial / DNA Miniprep Kit, Zymo Research). The 16S rRNA gene was amplified by PCR using universal bacterial primers 27F (5’AGAAGCGTAATTCAGGCAGTAC-3’) and 1492R (5’TACGGTACCTTGTTACGACTT-3’). The PCR product was purified by a QIA rapid PCR purification kit (Qiagen) and sequenced using the ABI PRISM model 3700 of the automatic DNA sequencer and the Big Dye Terminator (Applied Biosystems) cyclic sequence set. The 16S rRNA gene sequences of the isolates were searched for similarity to the NCBI BLAST sequences Altschul et al. (1990) and EzBio Cloud Kim et al. (2012) to identify the closest taxa. Phylogenetic and molecular evolution analyzes were performed using MEGA version 6 (references). The sequence sequence of 16s rRNA was transferred to the GENE BANK database.
Statistical analysis
The data was entered in the Microsoft® Excel 2007 spreadsheet.
Results
A liquid medium was used to check the depth of color, activity of Remazol Blue RGB and decolorization by bacterial isolates. Based on the relative ability to decolorize the various isolates, three of the most potent bacterial isolates (1, 2 and 3) with more than 90% decolorizing activity (data not shown) were preferred for further experiments.
Biodecolorization of remazol blue RGB by selected bacterial isolates
Microbial decolorization of Remazol Blue RGB with the most promising bacterial isolates (1, 2 and 3) was confirmed by another experiment in the liquid medium over a significant time frame (Figure 1). It was found that significant bacterial isolates had the proper ability to eliminate Remazol Blue RGB in actively growing cultures. The most suitable bacterial isolate for Remazol Blue RGB discoloration colorless was the 1st isolate with 98% degradation efficiency at 10 hours of incubation, while the remaining isolates showed the highest discoloration at 16h. Isolate 2 was the second highly qualified bacterial isolate and removed the Remazol Blue RGB color by up to 94% in 16 hours. Similarly, isolate 3 had a coloring potential of 86%
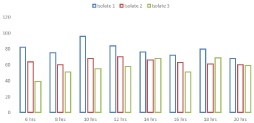
Figure 1: Biodecolorization of Remazol Blue RGB.
Factors affecting biodecolorization of remazol blue RGB in liquid medium
The potential of selected isolates (1, 2 and 3) was further investigated to optimize the various incubation /surrounding circumstances to remove the azo dye in the liquid medium. It was evident (Figure 2a,b,c) that the Remazol Blue RGB azo dye bleached through piercing to 100 mg of L-1 substrate concentration and the maximum removal was observed at 100 mg L-1 substrate concentration. Then there was a slow but certain decrease in azodye bleaching. Isolate 1 was the strongest azo color removal strain with more or less complete color removal, i.e. 100% decolorization at 100 mg L-1 and minimal decolorization was recorded at 50 mg L-1, whereas after substrate concentration 100 mg L-1, again Isolation 1 shows a decreasing trend. Isolation 2 was second in rank with 90% bleaching at 100 mg L-1. However, isolate 3 exhibited different trends from other isolates; indicates an increase in bleaching to up to 200 mg L-1 (82%).

Figure 2: A: Bioremediation of Remazol Blue RGB by isolate 1. B:
Bioremediation of Remazol Blue RGB by isolate 2. C: Bioremediation of
Remazol Blue RGB by isolate 3.
Carbon source effect
The particular effects of significant carbon sources such as maltose, mannitol, glucose and yeast extract have been evaluated for decolorization of Remazol Blue RGB bacterial isolates (Figure 3). It was found that in all selected strains (85-98%) the highest decolorization was carried out with 5% yeast extract, which was followed by glucose, in which decolorization occurred in the range of 20 to 25%. However, the least discoloration was observed for mannitol (10-15%) and maltose (up to 18%).
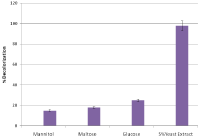
Figure 3: Effect of different Sources of carbon on decolorization of Remazol
Blue RGB.
To study pH optimization, the pH ranged from 5 to 9 and all selected isolates were inoculated there on (Figure 4). We start with a pH increase of 5 to 7, discoloration improves and the highest discoloration occurred at pH 7.5. Similarly, it had an auxiliary rise in pH from 7 to 9 a pessimistic result of decolorizing the mixture isolates. The highest decolorization was observed in the isolate 1 (98%) at pH 7.5, although the least discoloration occurs at pH 9. Comparable tendencies in the remaining isolates 2 and 3 were tested at pH 7.5. Overall, it was noted that all bacteria the isolates showed the most favorable staining from pH 5 to 7.5.
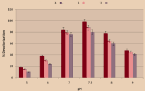
Figure 4: Effect of pH.
Incubation temperature effect
Five levels (25, 30, 35, 40 and 45°C) of the temperatures used in the evaluation of the optimal biodecolorization of Remazol Blue RGB by selected bacterial isolates. It is obvious (Figure 5) when the temperature has been raised from 25 to 35°C, there has been an inconsistent trend of discoloration due to different isolates. Isolate 1 showed a gradual increase in decolorization while isolate 3 shows a maximum bleaching at 25°C. The other two bacterial isolates (1 and 2) with a gradual rise from 25 to 35°C exhibited a maximum bleaching at 35°C. 35°C to 45°C, there was a sharp decrease in decolorization capacity in all isolates. The most bleached substance was detected by isolate 1 (98%) at 35°C followed by isolate 2 (94%) in a similar temperature. The smallest discoloration was observed at 45°C for all selected isolates.
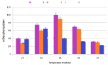
Figure 5: Effect of Incubation Temperature.
Identification
Among three selected bacterial isolates, isolate 1exhibited maximum decolorizing potential against azo dyes, so it was
identified by 16s rRNA gene sequencing approach. Sequence analysis of 16s rRNAgene showed that isolate 1 had 98.89% similarity with the genus bacillus. Phylogeny tree (Figure 6) based on Mega 6 indicated that isolate 1 was a Bacillus strain.

Figure 6: Phylogenetic tree derived from 16s rRNA gene sequence.
Discussion
Industrial waste water is very unstable and fluctuates repeatedly in a sufficient range depending on practical exercises. Most South Asian countries have severe environmental problems caused by ruined industrialization development. This observable fact is very widespread polluting industries such as paper, pulp, textiles, dyeing, leather, cocoa processing, and sugar production flourishes as clusters. Along with these industries, the textile industry is a huge industry water patrons, as well as wastewater producers. The waste water discharged by this sector has led to slight pollution groundwater and soil and, where appropriate, has an impact on the livelihood of the poor. The textile industry is one of the largest consumers of water and is considered the most polluting between all industries considering both volume and the composition of runoff on a global scale. It is a complex and highly variable mixture of many pollutants, from inorganic compounds, and elements for polymers and organic products. It produces a persistent color associated with organic load leading to a disruption of the overall ecological/symbiotic equilibrium of water inflow. Dyes with significant visibility in recipients can significantly affect photosynthetic activity in the aquatic environment due to reduce light penetration and may also be toxic some aquatic life caused by metals, chlorides, etc. associated with dyes or dyeing. The reason for reducing bleaching under shaking conditions could be the competition of oxygen and dye compounds for reduced electron carriers under aerobic conditions. The major part of the waste water contains azo dyes which, are increasingly used in industries due to their ease and cost-effective in synthesis compared to natural dyes. Relative efficacy of bacteria isolated for decolorization Remazol Blue RGB clearly indicates that they can be effective used to remove Remazol Blue RGB from textile waste water contaminated. Azoreductase is considered a key enzyme expressed in bacterial azo dyes, which degrade and catalyze reductive cleavage of the azo bond. Azoreductase activity was found in a number of bacterial species recently, such as Staphylococcus aureus, Shewanella putrefaciens, Shewanella and Pseudomonas spp. It was noted that increasing the substrate concentration of its best possible level had a pessimistic effect on decolourisation capacity of isolated isolates. Surveys with a strong dye concentrations in other experiments are also reported higher pure paint removal effects at lower dye concentrations. Reduce the ability to remove colors on a high surface the concentration can be due to the toxicity of the dye and what contaminants. On the aromatic ring of the azo dye structure one or more more sulfonic acid groups are generally approved, which can be active as detergents that inhibit the growth of microbes. Further explanation of toxicity at elevated concentration may be due to the presence of heavy metals or metal complexes dyes and / or the presence of unhydrolyzed reactive groups, which can inhibit the growth of bacteria. Similarly, diminution in color removal at stumpy concentration of the substrate might be due to the shrink in enzyme capability to be acquainted with the substrate proficiently. While yeast was experimented with different carbon sources the extract confirmed that it is the best of the tested carbon source. Our results were consistent with the research carried out by Guo et al., in which bacterial strains grew well and absolutely bleached K-2BP, where there is either peptone or yeast extract was contemporary in the medium; however, sucrose, glycerol, glucose, starch and lactose resulted in poorer rates of growth and decolorization of these dyes. Supplementary studies also accounted the utmost color removal of azo dyes in the presence of yeast extract by bacteria [29-31]. In case of variable pH, decolorization was on upper side at pH 7.5. Percentage of decolorization of Remazol Blue RGB from Bacillus spp. the strain under static conditions was 95% during 24 h of incubation, which was similar to the study but with the 35 h incubation period [32]. In another study carried out with Pseudomonas putida, P. fluorescence, Bacillus cereus and Stentrophomonas acidaminiphila to reveal Acid Red 88, 35%, 31%, 40% and 50% [33]. In aerobic conditions, azo dyes are generally resistant to bacterial attack [33]. The optimal pH for complete decoloration of Remazol Blue RGB was 8, which is slightly consistent with Consortium sp. Malachite green staining at pH 9 [34] and Klebsiella pneumonia RS-13, which completely degraded Methyl Red in pH 6 to 8 [34] While decolourisation decreased at alkaline pH the efficacy of all isolates tested was observed. So from this neutral pH could be closed in this research permanent bacterial movement to remove Remazol Blue RGB in a liquid medium. The temperature is even more incredible an important parameter for anaerobic wastewater treatment. The selected isolates were mesophilic bacteria because they all showed a better coloration in the temperature range of 25°C to 35°C. Similar results are also reported by Prasad et al., [35]. The mesophilic range is traditionally used because it is he generally believed that maintaining a high temperature would be uneconomic, while degradation within the psychrophilic range is too slow. Overall, one of the selected isolates of the bacteria was able to completely remove the color of the dye in 18 hours. However, these isolates should be tested on a large scale to explore their potential for bioremediation of contaminated dyes waste water.
Conclusion
The current study states that the three selected isolates it can be lucratively used to decolour Remazol Blue RGB dye. The cultures exhibited maximum decolorability at pH between 7.5 for all three isolates and 35°C for 1, 2 and 25°C for 3. Moreover, 5g/l of yeast extract was found to be optimal bleaching. In conclusion, isolate1 was found as one of most effective among others for bioremediation of Remazol Blue RGB,which can be further studied for bioremediation of dye polluted water including the degradation rate of azo dyes other than the selected blue dye through the application Bio augmentation.
References
- Abadulla E, Tzanov T, Costa S, Robra K, Cavaco A, Gubitz G. Decolorization and detoxification of textiles dyes with Laccase from from trametes hirsute. Appl Environ Microbial. 2000; 66: 3357-3362.
- Acuner E, Dilek FB. Treatment of tectilon yellow 2G by Chlorella vulgaris. Process Biochemistry. 2004; 39: 623-631.
- Aksu Z, Donmez G. A comparative study on the biosorption characteristics of some yeasts for Remozal Blue reactive dye, Chemosphere. 2003; 50: 1075-1083.
- Ambrosio ST, Takaki GMC. Decolorization of Reactive Azo dyes by Cunninghamella elegans UCP 542 under Co-metabolic conditions. Bioresour. Technol. 2004; 91: 69-75.
- Anjali P, Poonam S, Leela I. Bacterial decolorization and degradation of azo dyes. Int Biodet Biodegr. 2007; 59: 73-84.
- Saharan SB, Ranga P. Optimization of cultural conditions for decolourization of textile azo dyes by Bacillus subtilis spr42 under submerged fermentation. Advanced Biotechnology and Research. 2011; 2: 148-153.
- Bhatti HN, Akram N, Asgher M. Optimization of culture conditions for enhanced decolorization of Cibacron Red FN-2BL by Schizophyllum commune IBL-6, Appl Biochem Biotecnol. 2008; 149: 255-264.
- Chang JS, Chou C, Chen SY. Decolorization of azo dyes with immobilized Pseudomonas luteola, Process Biochemistry. 2001; 36: 757-763.
- Chang JS, Kuo TS. Kinetics of bacterial decolorization of azo dye with Escherichia coli NO3, Bioresource Technology. 2000; 75: 107-111.
- Chen KC, Wu JY, Liou DJ, Hwang SCJ. Decolorization of the textile azo dyes by newly isolated bacterial strains. J Biotechnol. 2003; 101: 57-68.
- Chen KC, Huang WT, Wu JY, Houng JY. Microbial decolorization of azo dyes by Proteus mirabilis. Journal of Industrial Microbiology & Biotechnology. 1999; 23: 686-690.
- Chen B, Zhang M, Chang C, Ding Y, Chen W, Hsueh C. Deciphering azo dye decolorization characteristics by indigenous Proteus hauseri: Chemical structure. J Thailand Inst. Chemical Engin. 2011; 42: 327-333.
- Chen B, Zhang M, Chang C, Ding Y, Chen W, Hsueh C. Deciphering azo dye decolorization characteristics by indigenous Proteus hauseri: Chemical structure. J Thailand Inst. Chemical Engin. 2011; 42: 327-333.
- Chen BY, Zhang MM, Chang CT, Ding YT, Lin KL, Chiou CS, et al. Assessment upon azo dye decolorization and bioelectricity generation by Proteus Hauseri. Bioresour Technol. 2010; 101: 4737-4741.
- Chen KC, Wu JY, Liou DJ, Hwang SJ. Decolorization of textile dyes by newly isolated bacterial strains. J Biotechnol. 2003; 101: 57-68.
- Chung KT, Fulk GE, Egan M. Reduction of Azo dyes by intestinal anaerobes. Appl Environ Microbiol. 1978; 35: 558-562.
- De-Bashan LE, Moreno M, Hernandez JP, Bashan Y. Removal of ammonium and phosphorus ions from synthetic wastewater by the microalgae Chlorella vulgaris coimmobilized in alginate beads with the microalgae growth- promoting bacterium Azospirillum brasilense. Water Research. 2012; 36: 2941-2948.
- Fournier D, Halasz A, Thiboutot S, Ampleman G, Dominic M, Hawari J. Biodegradation of octahydro- 1, 3, 5, 7- tetranitro-1, 3, 5, 7-tetrazocine (HMX) by Phnerochaete chrysosporium, New insight into the degradation pathway. Environmental Science and Technology. 2004; 38: 4130-4133.
- Fu Y, Viraraghavan T. Dye biosorption sites in Aspergillus niger, Bioresource Technology. 2002; 82: 139-145.
- Guo J, Zhou J, Wang D, Tian C, Wang PM, Uddin S. A novel moderately halophilic bacterium for decolorizing azo dye under high salt condition. Biodegradation. 2008; 19: 15-19.
- Gupta VK, Mittal A, Krishnan L, Gajbe V. Adsorption kinetics and column operations for the removal and recovery of malachite green from wastewater using ash. Separation and Purification Technology. 2004; 40: 87-96.
- Gupta VK, Rastogi A, Saini VK, Jain N. Biosorption of copper (II) from aqueous solutions by Spirogyra species. Journal of Colloid and Interface Science. 2006; 296: 59-63.
- Hsueh C, Chen B, Yen C. Understanding effects of chemical structure on azo dye decolorization characteristics by Aeromonashydrophila. J Haz Mat. 2009; 167: 995-1001.
- Hsueh CC, Chen BY. Comparative study on reaction selectivity of azo dye decolorization by Pseudomonas luteola. J Haz Mat. 2007; 141: 842-849.
- Hu TL. Degradation of azo dye RP2B by Pseudomonas luteola. Water Sci Technol. 1998; 38: 299-306.
- Jiunkins R. Pretreatment of textile waste water. Proc. 37th Industrial waste Conference Purdue Uni. Lafayette, Ind. 1982; 37-139.
- Kapdan IK, Oztekin R. Decolorization of textile dyestuff Reactive Orange 16 in fed-batch reactor under anaerobic condition. Enz Microbiol Technol. 2003; 33: 231-235.
- Khalid A, Arshad M, Crowley DE. Accelerated decolorization of structurally different azo dyes by newly isolated bacterial strains. Appl Microbiol Biotechnol. 2008; 78: 361-369.
- Khalid A, Arshad M, Crowley DE. Decolorization of azo dyes by Shewanella sp. under saline conditions. Appl Microbiol Biotechnol. 2008; 79: 1053-1059.
- Kumar KV, Sivanesan S, Ramamurthi V. Adsorption of malachite green onto Pithophora sp., a fresh water algae: equilibrium and kinetic modeling, Process Biochemistry. 2005; 40: 2865-2872.
- Maier J, Kandelbauer A, Erlacher A, Cavaco-Paulo A, Gubitz GM. A new alkali thermostable azo reductase from Bacillus sp. Strain SF. Appl Env Microbiol. 2004; 70: 837-844.
- Moorthi PS, Selvam SP, Sasikalaveni A, Murugesan K, Kalaichelvan PT. Decolorization of textile dyes and their effluents using white rot fungi, African J Biotech. 2007; 6: 424-429.
- Pinherio HM, Touraud E, Tomas O. Aromatic amines from azo dye reduction: status review with emphasis on direct UV spectrophotometric detection in textile industry wastewater, Dyes and Pigments. 2004; 61: 121-139.
- Ponraj M, Gokila K, Vasudeo Zambare. Bacterial decolorization of textile dye- Orange 3R, International journal of advanced biotechnology and research ISSN 0976-2612. 2011; 2: 168-177.
- Prasad A, Venkata K, Rao B. Pysico chemical analysis of textile effluent and decolorization of textile azo dyes by Bacillus endophyticus strain VITABR 13. IIOAB J. 2011; 2: 55-62.