
Research Article
Austin J Microbiol. 2021; 6(1): 1029.
Research of Associated Resistance to Antimicrobial Agents of Extended Spectrum Beta-Lactamases Escherichia Coli and Klebsiella pneumoniae Strains Isolated in Senegal
Ngom B¹*, Diagne R², Wade SF¹, Diop TA¹, Sow AI³
1Université Amadou Mahtar MBOW, Dakar, Sénégal/ Ecole Supérieure des Sciences Agricoles et de l’alimentation, Sénégal
2Université Iba Der Thiam, Thiès, Sénégal
3Université Cheikh Anta Diop de Dakar, Sénégal
*Corresponding author: Babacar Ngom, Université Amadou Mahtar MBOW, Dakar, Sénégal
Received: April 23, 2021; Accepted: May 17, 2021; Published: May 24, 2021
Abstract
Introduction: Some strains of Escherichia coli and Klebsiella pneumoniae produce Extended Spectrum Beta-Lactamases (ESBL) may be responsible for various infections such as urinary infections. These Sick people are treated in the very serious cases by association antibiotics to class to betalactamins, aminosids and quinolons. But proliferation of multi-drug resistant strains involves decreasing therapeutic success. That’s why epidemiological study must be done in all laboratories of bacteriology.
Purpose: The aim of the study was to research the resistance phenotypes of our E. coli and K. pneumoniae ESBL strains compared to others families of antibiotics.
Material and methods: Thirty two (32) Extended Spectrum betalactamases E. coli and K. pneumoniae strains isolated from either hospitalized patients or sick people who came for consultation were studied. Susceptibility to antimicrobial agents was determined using an antibiotic disk (Bio-Rad) diffusion method on Mueller-Hinton agar (Bio-Rad). The results were interpreted according to the Standards of the French Antibiogram Committee (CA-SFM).
Results: The study showed that most of these strains were multi-drug resistant. They were resistant to many beta-lactamines antibiotics. E. coli strains were also resistant at 70,34% to aminosids, at 96,72% to quinolons, at 58,3% to cotrimoxazol, at 26,1% to chloramphénicol and at 21,4% to colistin ; about K. pneumoniae, they were resistant at 72,6% to aminosids, at 88,95% to quinolons, at 86,7% to cotrimoxazol, at 44,4% to chloramphénicol and at 25% to colistin. But all these strains were sensitive at 100% to l’imipenem.
Keywords: E. coli, K. pneumoniae, ESBL, multi-drug resistant bacteria, Senegal
Introduction
The resistance of bacteria to antibiotics is now a “globalized” phenomenon, affecting all bacterial species of medical importance and all curent classes of antibiotics [1,2]. It is the result of the excessive dissemination of antibiotics, for many uses (human and veterinary medicine, breeding and agriculture). This is the case with beta-lactams, which are the mainstay of antibiotic therapy for enterobacterial infections [3]. In fact, some potentially pathogenic strains of this family, such as E. coli and K. pneumoniae, which produce extended spectrum betalactamases (ESBL) and are frequently isolated from urinary tract infections [4-6], can be resistant to many molecules of this class. The most worrying thing is that these strains, which spread rapidly both in hospitals and in the community, are often co-resistant to many other antibiotics such as those of the aminoglycoside and quinolone class, thus making the treatment of these very problematic infections [4,7,8].
Material and Methods
Strains
Sixty-four strains of which thirty-two E. coli and K. pneumoniae were the subject of this study. These strains were isolated from patients hospitalized in various departments of the Fann national university hospital or received in outpatient consultations. The pathological products in which these strains have been isolated are: urine, blood, pus and vaginal sample and bronchoalveolar fluid. These strains were identified according to their morphological, cultural and biochemical characteristics (Api 20E gallery -bioMérieux) and were all secretors of Extended Spectrum Beta-Lactamases (ESBL).
Antibiogram
The study of the sensitivity of strains to antibiotics (antibiogram) was carried out using the agar medium diffusion method according to the updated recommendations of the Antibiogram Committee of the French Society of Microbiology (http://www.sfm.asso.fr/). The ESBL screening was performed by placing, 3 cm center to center, discs of ceftazidime, ceftriaxone, cefotaxime, cefepime and/or aztreonam around an Amoxicillin/clavulanic acid disc. The presence of a “champagne cork” synergy between Amoxicillin/clavulanic acid and one of the aforementioned discs attests to the secretion of ESBL by the strain studied. Other antibiotics, both belonging to the beta-lactam family and not, have been tested. These are amoxillin, ticarcillin, piperacillin, cephalotin, cefoxitin, imipenem, chloramphenicol, nitroxoline, colistin, cotrimoxazol (sulfametoxazol/trimetopime) aminoglycosid (kanamicin), tobramicin, gentamicin, netilmicin and amikacin) and quinolones (nalidixic acid, norfloxacin, pefloxacin and ciprofloxacin).
Results
Distribution of strains by department - Results showed a predominance of strains of E. coli (Figure 1) and K. pneumoniae (Figure 2) in the infectious disease department with respective percentages of 53.13% and 37.5% of isolates (Table 1).
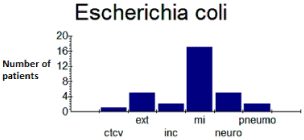
Figure 1: Distribution of E. coli by service.
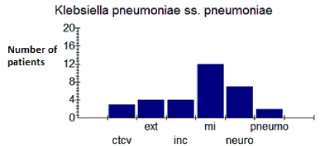
Figure 2: Distribution of K. pneumoniae strains according to the departments.
Species
Number of strains
ctcv
ext
inc
mi
neuro
pneumo
Escherichia coli
32
1
5
2
17
5
2
Klebsiella pneumoniae
32
3
4
4
12
7
2
Table 1: Distribution of strains according to services.
Distribution of strains according to samples
The majority of strains studied were isolated from urine. In fact, 84.37% of E. coli (Figure 3) and 56.25% of K. pneumoniae (Figure 4) came from the urine. Table 2 gives the distribution of the strains according to the samples.
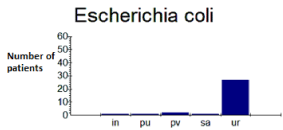
Figure 3: Distribution of E. coli according to pathological products.
In: Unknown; pu: Urine Sample; pv: Vaginal Sample; sa: Blood; ur: Urine
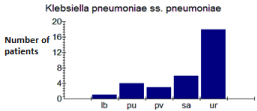
Figure 4: Distribution of K. pneumoniae according to pathological products.
Lb: Bronchoalveolar Fluid
Species
Number of strains
in
Lb
pu
pv
sa
ur
Escherichia coli
32
1
-
1
2
1
27
Klebsiella pneumoniae
32
-
1
4
3
6
18
Table 2: Distribution of strains according to samples.
Antibiogram results for E. coli
Table 3 gives the results for antibiograms in E. coli in percentages. Thus, high resistance of ESBL E. coli has been noted to penicillins, carboxypenicillins, ureidopenicillins, first and third generation cephalosporins and aztreonam, as well as to aminoglycosides (except amikacin) and quinolones. However, these strains remain very sensitive to imipenem (100%), cefoxitin (63.3%) and colistin (78.6%), amikacin (63.3%) and chloramphenicol (73.9%) (Figure 5).
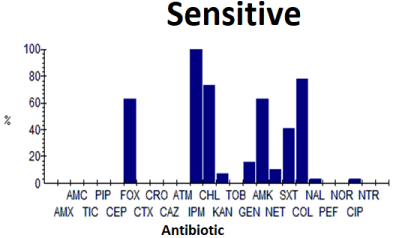
Figure 5: Antibiotic sensitivity of E. coli.
Codes
Antibiotic
Nbre
%R
%I
%S
AMX_ND25
Amoxicillin
31
100
0
0
PIP_ND75
Piperacillin
31
100
0
0
TIC_ND75
Ticarcillin
31
100
0
0
AMC_ND20
Amoxicillin/clavulanic Acid
32
71,9
28,1
0
CEP_ND30
Cefalothin
30
100
0
0
CAZ_ND30
Ceftazidime
29
100
0
0
CRO_ND30
Ceftriaxone
25
100
0
0
CTX_ND30
Cefotaxime
23
100
0
0
FOX_ND30
Cefoxitin
30
10
26,7
63,3
ATM_ND30
Aztreonam
32
100
0
0
IPM_ND10
Imipenem
31
0
0
100
AMK_ND30
Amikacin
30
13,3
23,3
63,3
GEN_ND15
Gentamicin
31
80,6
3,2
16,1
KAN_ND30
Kanamycin
26
92,3
0
7,7
NET_ND30
Netilmicin
29
65,5
24,1
10,3
TOB_ND10
Tobramycin
26
100
0
0
NAL_ND30
Nalidixic Acide
32
96,9
0
3,1
NTR_ND20
Nitroxolin
16
0
100
0
CIP_ND5
Ciprofloxacin
26
96,2
0
3,8
NOR_ND5
Norfloxacin
16
93,8
6,2
0
PEF_ND5
Pefloxacin
21
100
0
0
SXT_ND1.2
Trimethoprim/Sulfamethoxazol
12
58,3
0
41,7
COL_ND50
Colistin
28
21,4
0
78,6
CHL_ND30
Chloramphenicol
23
26,1
0
73,9
Table 3: Antibiogram results for E. coli.
Antibiogram results for K. pneumonia
The strains of K. pneumoniae studied were very resistant to penicillins, carboxypenicillins, ureidopenicillins, first and third generation cephalosporins and aztreonam, as well as aminoglycosides (except amikacin) and quinolones (Table 4). However, the latter were sensitive to varying degrees to imipenem (100%), cefoxitin (88.9%) and colistin (75%), amikacin (83.9%) and chloramphenicol. (55.6%) (Figure 6).
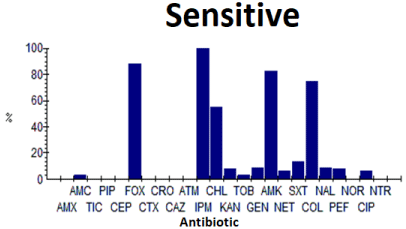
Figure 6: Antibiotic sensitivity of K. pneumoniae.
Codes
Antibiotiques
Nbre
%R
%I
%S
AMX_ND25
Amoxicillin
32
100
0
0
PIP_ND75
Piperacillin
32
100
0
0
TIC_ND75
Ticarcillin
29
100
0
0
AMC_ND20
Amoxicillin/Clavulanic Acid
31
61,3
35,5
3,2
CEP_ND30
Cefalothin
25
100
0
0
CAZ_ND30
Ceftazidim
29
100
0
0
CRO_ND30
Ceftriaxon
28
100
0
0
CTX_ND30
Céfotaxim
24
100
0
0
FOX_ND30
Céfoxitin
27
0
11,1
88,9
ATM_ND30
Aztreonam
24
100
0
0
IPM_ND10
Imipenem
31
0
0
100
AMK_ND30
Amikacin
31
12,9
3,2
83,9
GEN_ND15
Gentamicin
31
90,3
0
9,7
KAN_ND30
Kanamycin
24
79,2
12,5
8,3
NET_ND30
Netilmicin
32
84,4
9,4
6,2
TOB_ND10
Tobramycin
26
96,2
0
3,8
NAL_ND30
Nalidixic acid
32
84,4
6,2
9,4
NTR_ND20
Nitroxolin
10
0
100
0
CIP_ND5
Ciprofloxacin
32
93,8
0
6,2
NOR_ND5
Norfloxacin
20
95
5
0
PEF_ND5
Pefloxacin
23
82,6
8,7
8,7
SXT_ND1.2
Trimethoprim/Sulfamethoxazol
15
86,7
0
13,3
COL_ND50
Colistin
28
25
0
75
CHL_ND30
Chloramphenicol
27
44,4
0
55,6
Table 4: Antibiogram results for K. pneumoniae.
Discussion
The study showed the circulation of ESBL-producing E. coli and K. pneumoniae strains in both hospital and community settings. Many strains have been isolated from patients who came for an outpatient consultation.
Most of the strains came from the infectious diseases department, which could be justified by nosocomial infections with E. coli and K. pneumoniae if we know that the patients hospitalized in this department are most often immunocompromised.
Most of the strains came from urine. Studies have shown that these multidrug-resistant strains express numerous virulence factors responsible for their uropathogenicity [9-11].
The strains studied were very sensitive to impipenem (100%). A few of them (10% of E. coli) were resistant to cefoxitin whose activity is comparable to that of cephamycin. ESBL-secreting strains only must be sensitive to this molecule. Their resistance to cefoxitin is therefore the result of a resistance mechanism other than ESBL secretion. It seems to be caused by the production of a class C beta-lactamase. In fact, work carried out in several countries [12-14] had revealed the emergence of E. coli and K. pneumoniae strains resistant to cefoxitin thanks to the production of beta-lactamase (cephalosporinase) AmpC plasmid [15-17].
The ESBL strains studied showed a high rate of co-resistance to antibiotics. Indeed, several of these strains were also resistant to several antibiotics of different classes other than beta-lactams, in particular aminoglycosid and quinolones.
E. coli were 80.6%, 100%, 65.5% and 92.3% resistant to gentamicin, tobramicin, netilmicin and kanamicin, respectively and 96.9%, 100%, 96.2% and 93.8% respectively to nalidixic acid, pefloxacin, ciprofloxacin and norfloxacin. As for K. pneumoniae, they were also resistant to 90.3%, 96.2%, 84.4% and 79.2% respectively to gentamicin, tobramicin, netilmicin and kanamicin and to 84.4%, 82.6%, 93.8% and 95% respectively to nalidixic acid, pefloxacin, ciprofloxacin and norfloxacin. However, these strains had high sensitivities to amikacin, 63.3% for E. coli and 83.9% for K. pneumoniae.
They were also resistant to colistin, cotimoxazol and chloramphenicol to varying degrees. E. coli were 21.4% resistant to colistin, 58.3% to cotrimoxazol and 26.1% to chloramphenicol and K. pneumoniae 25% to colistin, 86.7% to cotrimoxazol and 44.4% with chloramphenicol. These results are a perfect illustration of the multiresistant character of ESBL-producing enterobacteriaceae strains as described around the world [18], which complicated the treatment of the pathologies caused by the latter and necessitated a national surveillance of resistance. Antibiotics of these strains in order to prevent their spread and also that in the future we are faced with situations of therapeutic impasses.
Conclusion
The results of the study showed that the ESBL-secreting strains studied very often exhibited co-resistance against other families of antibiotics. They also show the spread of these strains both in hospitals and in the community. Their emergence requires very broad rigorous epidemiological surveillance of resistance to antibiotics in order to prevent their dissemination and that in the future we may be faced with situations of therapeutic impasses.
References
- Levy SB, O’Brien TF. Antimicrobial alerts and implications. Clinical Infect Dis. 2005; 41: 219-220.
- Bonnedahl J, Drobni M, Gauthier-Clerc M, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulles in the south of France. PLoS One. 2009; e5958.
- Slama TG. Gram-negative antibiotic resistance: there is a price to pay. Crit Care. 2008; 12: S4.
- Bonnedahl J, Drobni M, Gauthier-Clerc M, et al. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulles in the south of France. PLoS One. 209; e5958.
- Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing Extended-Spectrum Beta-Lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005; 56: 52-59.
- Zahar JR, Lortholary 0, Martin C, Potel P, Nordmann P. Addressing the challenge of extended-spectrum β-lactamases. Curr Opin Investig Drugs. 2009; 10; 172-180.
- Paterson DL, Bonomo R. Extended-spectrum β-lactamases: a clinical update. Clin Microbial Rev. 2005; 18: 657-686.
- Pitout JD, Laupland KB. Extended-spectrum β-lactamase producing Enterobacteriaceae: an emerging public health concern. Lancet Infect Dis. 2008; 8: 159-166.
- Mulvey MA, Schilling JD, Martinez JJ, Hultgren S. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc nat Acad Sci. 2000; 97: 8829-8835.
- Mulvey MA. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 2002; 4: 257-271.
- Johnson JR, Kuskowski MA, Gajewski A, et al. Extended virulence genotypes and phylogenetic background of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J infect Dis. 2005; 191: 46-50.
- Bauernfeind A, Hohl P, Schneider I, Jungwirth R, Frei R. Escherichia coli producing a cephamycinase (CMY-2) from a patient from the Libyan-Tunisian border region. Clin Microbiol Infect. 1998; 4: 168-170.
- Doi Y, Shibata N, Shibayama K, Kamachi K, Kurokawa H, Yokoyama K, et al. Characterization of a novel plasmid-mediated cephalosporinase (CMY-9) and its genetic environment in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2002; 46: 2427-2434.
- Manchanda V, Singh NP. Occurrence and detection of AmpC betalactamases among Gram-negative clinical isolates using a modified threedimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003; 51: 415-418.
- Philippon A, Arlet G, and Jacoby GA. Plasmid-determined AmpC type β-lactamases. Antimicrob. Agents Chemother. 2002; 46: 1-11.
- Odeh R, Kelkar S, Hujer AM, Bonomo RA, Schreckenberger PC, Quinn JP. Broad resistance due to plasmid-mediated AmpC betalactamases in clinical isolates of Escherichia coli. Clin Infect Dis. 2002; 35: 140-145.
- Wong-Beringer A, Hindler J, Loeloff M, Queenan AM, Lee N, Pegues DA, et al. Molecular correlation for the treatment outcomes in bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae with reduced susceptibility to ceftazidime. Clin Infect Dis. 2002; 34: 135-146.
- Jacoby S. Properties of plasmid responsible for production of extended spectrum bêta-lactamases antimicrob. Agent chemother. 1991; 35: 164-169.