
Research Article
Austin J Microbiol. 2021; 6(1): 1030.
Enhanced CAUTI Risk due to Strong Biofilm Forming MDR Bacteria in Indwelling Urinary Catheters
Gopinathan H, Johny TK and Bhat SG*
Department of Biotechnology, Cochin University of Science and Technology, Kerala, India
*Corresponding author: Sarita G Bhat, Department of Biotechnology, Cochin University of Science & Technology, Kochi-682022, Kerala, India
Received: May 20, 2021; Accepted: June 15, 2021; Published: June 22, 2021
Abstract
The achievement of modern medicine is due to development and extensive use of indwelling biomedical devices like urinary catheters, heart valves, Vascular bypass grafts, ocular lenses and artificial joints, among others. Untreated nosocomial infections due to urinary catheter- biofilms pose great health risk to patients. This study focuses on the isolation, molecular identification, antibiotic susceptibility profiling and physicochemical characterization of strong biofilm producers from indwelling urinary catheters. Out of 34 isolates 19 strong biofilm producers were segregated using Microtitre plate and Congo red agar methods. Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli, Morganella morganii and Enterococcus faecalis, which are common causative agents of Catheter-Associated Urinary Tract Infection (CA-UTI) were identified by molecular characterization and phylogenetic analyses. All strong biofilm formers were multi drug resistant by modified Kirby- Bauer method and Multiple Antibiotic Resistance (MAR) index was also calculated Further physicochemical characterization included hydrophobicity and autoaggregation assays. All the strong biofilm producers exhibited multiple antibiotic resistance. More than 60 per cent of the selected strains were strongly hydrophobic. No significant connection between autoaggregation and hydrophobicity was observed. All the characteristics of these strains including biofilm formation, multiple antibiotic resistance, hydrophobicity and auto aggregation abilities made them strong candidates for CA-UTI.
Keywords: CAUTI; Biofilm; Nosocomial infection; Multiple antibiotic/drug resistance; Hydrophobicity
Abbreviations
CAUTI: Catheter-Associated Urinary Tract Infection; MAR: Multiple Antibiotic Resistance; TSB: Tryptone Soy Broth; PBS: Phosphate-Buffered Saline; OD: Optical Density; PCR: Polymerase Chain Reaction; BLAST: Basic Local Alignment Search Tool; NCBI: National Centre for Biotechnology Information; MEGA: Molecular Evolutionary Genetics Analysis
Isolate
Organism
Genbank accession number
BTTLA2
Klebsiella pneumoniae
MG957112
BTTDC5
Klebsiella pneumoniae
MG648415
BTTDD4
Klebsiella pneumoniae
MG648421
BTTDD3
Pseudomonas aeruginosa
MG648420
BTTDD5
Escherichia coli
MG648422
BTTDD6
Morganella morganii
MG648423
Table 1: Identity of the selected strong biofilm producing multiple antibiotic resistant bacteria.
Introduction
Biotic as well as abiotic surfaces accommodate microbial biofilms. Microorganisms in biofilms exert direct or indirect influences on human life in terms of diseases, piped water, food and contact surfaces contamination in the healthcare sector, biofilms critically contaminate indwelling medical devices such as a prosthetic heart valve, pacemakers, central venous catheter, urinary catheter, contact lenses, and intra-uterine devices, leading to causing persistent and deadly infections [1].
Urinary catheters are silicone or latex rubber tubes used to eliminate urine from the urinary compartments and are the second most commonly used indwelling device by people with urinary retention and incontinence in the last decade [2]. This has consequently increased incidence of nosocomial infections during the first 10-14 days of catheterization in patients by over 40% [3].
Short-term catheterization promotes intensive biofilm formation on the external surface. But biofilm formation on the inner lumen of the catheter ensues after long-term catheterization (30 days or more) [4]. Biofilm-forming strains of Escherichia coli, Pseudomonas aeruginosa, Enterococcus, Proteus mirabilis and Klebsiella pneumoniae commonly contaminate urinary catheter and develop biofilms [5].
Autoaggregation and surface hydrophobicity are two independent traits that offer indirect methods for evaluating the adhesion by bacteria and is corroborated by numerous studies [6]. Bacterial adhesion initiated by the formation of a protein film due to deposition of urinary components on catheter surfaces, instigates biofilm formation [7,8]. Antimicrobial therapy is not necessitated if the catheter is removed or replaced from patients with asymptomatic bacteriuria, however. symptomatic patients do A worrisome feature of biofilm-based infections [9,10] is the high antibiotic resistance of bacterial and fungal cells within the biofilm [11,12]. Biofilms represent an ideal niche for plasmid exchange among bacteria. The conjugation frequency appears to be higher in bacteria growing in the sessile mode than in the planktonic mode. As some plasmids contain genes coding for multidrug resistance, microbial biofilms provide a suitable environment to amplify both naturally occurring and induced antibiotic resistance phenomena. Therefore, multiple drug resistance of urinary pathogens to common antibiotics is a topic of paramount concern.
Identification of bacteria causing CAUTI and their antibiotic sensitivity pattern is crucial for treatment as well as prevention of CAUTI. This study therefore aims to isolate and characterize bacteria from long duration indwelling urinary catheter to ascertain the level of risks imposed by them, enabling new treatment modalities for CAUTI.
Materials and Methods
Isolation and identification of bacteria from urinary catheters
Four foley urinary catheters were collected from patients catheterized for 21 days and placed aseptically in sterile containers. They are named as BTTLA, BTTLB, BTTDC and BTTDD. The catheters were analyzed using standard plate assay. Sections 1-2 cm and 3-4 cm from the catheter tip were cut, washed with sterile distilled water and suspended in phosphate-buffered saline (10 mL) in sterile test tubes. The colonizing biofilms were disrupted by vortexing for 2 minutes. 0.1 mL of sample serially diluted in (1:10) sterile distilled water was plated on Hi-Chrome UTI Agar (Hi-Media) plates. The plates were incubated for 18-24 h and colonies were isolated and streaked on slants.
Qualitative analysis for the biofilm producers by Congo red agar assay
Qualitative analysis for biofilm producers was done by Congo red assay method [13]. The isolates from UTI agar were spotted and incubated at 37°C for 24 h. Black colonies with a dry crystalline consistency indicate biofilm are observed. The experiment was performed in triplicate. although the method is stated as reliable,there is a possibility of false-positive results [14] and hence need to be confirmed using microtiter plate assay.
Quantitative analysis of biofilm formation
The bacterial isolates from urinary catheters were tested for their biofilm-forming ability using microtiter plate assay [15]. Organisms isolated from UTI agar plates were inoculated in 3 mL of tryptone soy broth and incubated at 37°C for 24 h. 230 μL of Tryptone Soy Broth (TSB) (HiMedia, Mumbai, India) was added to the wells of a sterile 96 well polystyrene microtiter plate. 20 μL bacterial cultures (OD600 =1) were added into each well separately. The experiment was performed in triplicate. The plate was incubated aerobically for 24 h at 37°C. Uninoculated sterile broth served as negative control. After incubation, the contents of each well were poured off. The wells were washed 3 times with phosphate buffer (0.01 M, pH 7.2) to eliminate free-floating bacteria. Biofilm produced by bacteria adherent to the wells were fixed with methanol. The plates were decanted after 15 minutes, air-dried and stained using 1% crystal violet for 5 min. The excess stain was removed by rinsing under running tap water, followed by air drying. The dye bound to adherent cells was extracted with 33% (v/v) glacial acetic acid and the absorbance was measured at 570 nm using a UV-VIS spectrophotometer (Shimadzu, Japan). Based on the absorbance (A570), they were classified as follows.
OD = ODC - Non-adherent
ODC < OD ≤ 2 X ODC - Weakly adherent
2 X ODC < OD = 4X ODC - Moderately adherent
4 X ODC < OD - Strongly adherent
where cut-off absorbance ODc was the mean absorbance of the negative control. All data from biofilm quantitative assays were expressed as mean ± SD with each assay conducted in triplicates.
Molecular characterization of biofilm producers
A single colony of the strain from UTI agar (Hi-Media) was picked using a sterile tip was re-suspended in 20 μL of nuclease-free water and vortexed for 30s. This was then subjected to heat shock at 95°C for 15 min in a thermo-cycler. Then 1 μL of the lysate was used as a template for PCR amplification of the 16S rDNA using universal primer pair [16]. The sequence identity was determined by comparing the 16S rDNA sequence with the sequences available in the NCBI nucleotide database using BLAST (Basic Local Alignment Search Tool) algorithm [17]. A phylogenetic tree was also constructed for the biofilm producers by the neighbour-joining method [18] using the MEGA 7 software [19].
Antibiotic sensitivity tests
All strong biofilm producers were screened for antibiotic sensitivity according to the Kirby- Bauer method [20], with 12 antibiotics (Hi-Media, Mumbai, India) belonging to different classes. It included the aminopenicillin antibiotic ampicillin (5 μg/ disc), macrolide antibiotic azithromycin (15 μg/disc), cephalosporin antibiotic cefixime (5 μg/disc), second-generation cephalosporin cefuroxime (30 μg/disc), semisynthetic, broad-spectrum antibiotic chloramphenicol (30μg/disc), Fluoroquinolone antibiotic ciprofloxacin (5μg/disc), Aminoglycoside antibiotic gentamicin (10μg/disc), synthetic quinolone antibiotic nalidixic acid (30μg/disc), fluoroquinolone antibiotic norfloxacin (5 μg/disc), broad-spectrum polyketide antibiotic tetracycline (30 μg/disc), and antifolate antibiotic trimethoprim (5 μg/disc). The results were interpreted as per manufacturer’s instructions. (HiMedia, India) [21].
MAR (Multiple Antibiotic Resistance) indexing
MAR index was calculated using the formula a/b, where ‘ a’ is the number of antibiotics to which the isolate was resistant, and ‘b’ is the number of antibiotics to which the isolate was exposed [22].
Surface hydrophobicity assay
Surface hydrophobicity assays were performed according to Kos et al. (2003) [23]. Bacteria were harvested in the stationary phase by centrifugation at 5000 x g for 15 min, washed twice and re-suspended in PBS (pH 7.4) to obtain 0.1 OD at 600nm. The absorbance of the cell suspension was measured at 600 nm (OD1). Aliquots of 1.2 mL sample were placed in duplicate test tubes and 0.5 mL of p-xylene was added to each tube. After a 10 min pre-incubation at 30°C, the two-phase system was mixed by vortexing for 60s. The aqueous phase was removed after 20 min of incubation at room temperature, and its absorbance at 600 nm (OD2) was measured. The percentage of hydrophobicity was calculated as OD1-OD2/OD1*100. Strains were classified as non-hydrophobic (<20%), moderately hydrophobic (20– 50%), and strongly hydrophobic (>50%) [24].
Autoaggregation Assay
Autoaggregation assays were executed according to Del Re et al. 2000) with modifications. Bacteria were grown for 18 h at 37°C in Nutrient brothand harvested by centrifugation at 5000 x g for 15 min, washed twice and suspended in phosphate-buffered saline (PBS) to get OD600 =1. Cell suspensions (4 mL) were vortexed for 10 s. Autoaggregation was determined during 5 h of incubation at room temperature. Every hour 0.1 mL of the upper suspension was transferred to another tube with 3.9 mL of PBS and the absorbance (A) was measured at 600 nm. The autoaggregation percentage is expressed as: 1- (At/A0) x 100, where At represents the absorbance at time t = 1, 2, 3, 4 or 5 h and A0 the absorbance at t = 0.
Results and Discussion
Screening for bacterial pathogens from urinary catheters
Four (04) patients within the age range of 61-75 years, with indwelling urinary catheters were screened during the study period. The duration of the catheterization was 21 days.
Qualitative analysis for the biofilm producers by Congo red assay
In the qualitative assay with Congo red agar plate, only 18 of the 34 isolates produced black colonies with crystalline appearance indicative of strong biofilm formers, 8 were moderate biofilm producers and the remaining 8 showed weak biofilm-production.
Quantification of biofilm-producing pathogens by microtitre plate assay
The quantitative assay showed a slight difference from that of the qualitative assay, with 55.8% (n=19) proved as strong biofilm formers, 14.70% (n=5) as moderate producers while 29.41% (n=10) were weak producers. Biofilm quantification by the 19 strong biofilm producers by Microtitre Plate assay is indicated in Figure 1 and the categorization and comparison of biofilm producers by Congo Red Agar method and Microtitre plate assay method are as depicted in Figure 2. Out of the nineteen strong producers, maximum biofilm formation was by strain BTTDD4 Klebsiella pneumoniae. All strong biofilm formers were further identified using 16S rDNA sequence analysis.
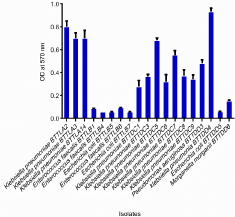
Figure 1: Quantification of Biofilm production by 19 strong biofilm producers.
Quantification of Biofilm production by 19 strong biofilm producers by
Microtitre plate method -ODC -0.006, 2ODc - 0.012, 4ODC-0.024.Values
<0.006 do not indicate biofilm producers, Values >0.006 but less than 0.012
indicate weak biofilm production. Values between 0.012 and 0.024 imply
moderate biofilm production and. values >0.024 are indicative of strong
biofilm production. Here, cut off absorbance ODc was the mean absorbance
of the negative control.
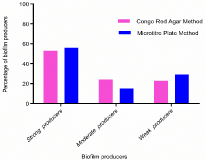
Figure 2: Comparison of biofilm producers by Congo Red agar method and
Microtitre plate method.
Molecular characterization of strong biofilm producers using 16S rDNA sequence analysis
Colony PCR based 16S rDNA amplification of the nineteen isolates and sequence analysis thereafter was used for molecular characterization of the biofilm producers. Following BLAST, the identity of the biofilm producers was determined. The sequence data were submitted to the NCBI GenBank and accession numbers were obtained.
The 16S rDNA sequence analysis enabled identification of the 19 strong biofilm formers belonging to the genera Klebsiella, Enterococcus, Escherichia, Pseudomonas, and Morganella. Eleven Klebsiella sp., three Enterococcus sp., three E. coli, one each of Morganella and Pseudomonas sp. were identified using 16S rDNA analysis.In the current study, Gram-negative bacteria were more dominant (84.21 %) than Gram-positive bacteria (15.78%). Klebsiella pneumoniae (57.8 %) constituted the predominant bacteria, followed by Enterococcus faecalis (15.7 %), E. coli (15.7%), Pseudomonas aeruginosa and Morganella morganii (5 .2%) each.
Phylogenetic analysis of the biofilm-producing strains obtained in the study was performed to understand their interrelatedness and is depicted in Figure 3. The Neighbour-Joining method was used to infer the evolutionary history [18]. The optimal tree with the sum of branch length = 0.35206479 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. 19 nucleotide sequences were involved in the analysis. All positions containing gaps and missing data were eliminated. There were a total of 387 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [19].

Figure 3: Phylogenetic analysis of the biofilm-producing strains.
Antibiogram of the strong biofilm producers
Antibiotic sensitivity profile of the strong biofilm producers is as given in Figure 4. It was observed that all biofilm producers were resistant to cefixime, cefuroxime and nalidixic acid followed by ampicillin, ciprofloxacin, norfloxacin, tetracycline and trimethoprim. A higher percentage of biofilm producers showed resistance (Resistance, Intermediate resistance) to azithromycin. However, the isolates showed a greater sensitivity towards chloramphenicol and gentamycin.
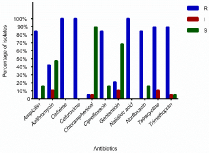
Figure 4: The antibiogram (%) of strong biofilm producers.
MAR Index
All organisms showed high MAR indices implying high antibiotic resistance and strong biofilm producing nature. The MAR index values are as shown in Figure 5. Out of the nineteen strong biofilm producers, six urinary catheter strains were selected based on their strong biofilm production and high antibiotic resistance profile for further studies.
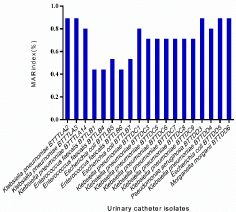
Figure 5: MAR index (%) of strong biofilm producers.
Surface hydrophobicity assay
Six selected urinary catheter strains were subjected to cell surface hydrophobicity and the results are depicted in Figure 6. Four strains, Klebsiella pneumoniae BTTLA2, Pseudomonas aeruginosa BTTDD3, Klebsiella pneumoniae BTTDD4 and Escherichia coli BTTDD5 were strongly hydrophobic and the other two strains were moderately hydrophobic. Hydrophobicity of these bacterial cells can increase the tendency of microorganisms to adhere to the hydrophobic surfaces of indwelling medical devices and biofilm formation [25].
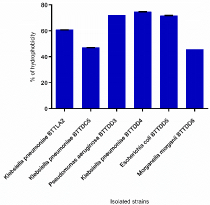
Figure 6: Surface hydrophobicity of six strong biofilm producers.
Auto aggregation Assay
Auto aggregation assay results of six selected urinary catheter isolates are as shown in Figure 7. Morganella morganii BTTDD6 possessed the highest autoaggregation capacity in comparison to the other strains, viz. Klebsiella pneumoniae BTTLA2, Klebsiella pneumoniae BTTDC5, Pseudomonas aeruginosa BTTDD3, Klebsiella pneumoniae BTTDD4 and Escherichia coli BTTDD5. Auto aggregation of all the selected strains increased with time.
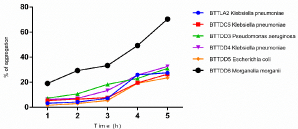
Figure 7: Auto aggregation ability of six strong biofilm producers.
Urinary catheters are medical devices made of tubular latex or silicone, used in both hospital and healthcare facilities. The most frequent complication associated with these devices is the development of biofilms on the inner or outer surfaces leading to Catheter-Associated Urinary Tract Infections (CAUTIs) [26–28], which is a serious health threat to patients if untreated. Bacteria associated with biofilm exhibit endurance to antibiotic therapy. So, the development of better catheter surfaces and improved treatment options requires an improved understanding of the uropathogens on the catheter and their antibiotic resistance profiles.
In this study, biofilm forming ability of 34 clinical strains selectively isolated from the urinary catheters from patients were evaluated using Congo red agar and Microtitre plate assay methods. Congo red agar plate assay is a phenotypic method to detect biofilm formation, that is fast, reproducible and which retains cell viability, permitting further analysis of these colonies. Spot inoculation gave better visualization and easier interpretations, especially for the biofilm-producing strains. The results are interpreted based on the colour of the colonies which ranged from red for non-biofilm– producing strains to black for biofilm-producing strains [29]. Black precipitated colonies were produced by the reaction of the strong slime with the media components, and the indicator Congo red. The intensity of colony colour is directly proportional to extent of biofilm production. Strong biofilm producers produce dark black coloured colonies. The decrease in strength of biofilm production reduces the intensity of the colony colour. The method cannot be stated as reliable since there is a possibility of false-positive results [14] and hence requires confirmation by quantitative assays such as the Microtitre plate biofilm assay.
Microtitre plate biofilm assay is a simple high-throughput method used to monitor microbial attachment to an abiotic surface. The results of the Microtitre plate biofilm assay method showed that most of the strains in the study formed biofilms on plastic surfaces. The strong biofilm-producing isolates like K. pneumoniae, P. aeruginosa, M. morganii, E. coli and E. faecalis were selectively isolated and identified from catheters. Members of the Family Enterobacteriaceae within class Gammaproteobacteria were most abundant and prevalent in this study. Klebsiella pneumoniae accounting for 57.8% of the total isolates was the most common pathogen among the strong biofilm producers isolated in the present study, with strain Klebsiella sp. (BTTD4). being the strongest biofilm producer. Klebsiella pneumoniae, an opportunistic pathogen, is a frequent cause of CAUTIs [30–33], being responsible for up to 10% of all nosocomial bacterial infections and causes UTIs, septicemia, or pneumonia in immune-compromised individuals [34–36]. Type 1 or type 3 fimbriae, capsule and the LPS are the virulence factors mostly involved in the ability of K. pneumoniae to flourish as biofilm [37]. Type 1 fimbriae mediate the adherence to many types of epithelial cells and may facilitate adherence of the bacteria to the bladder epithelium. Klebsiella pneumoniae requires type 3 fimbriae for biofilm formation on plastics and human extracellular matrix; As a result, they may facilitate the formation of treatment-resistant biofilm on indwelling plastic devices, such as catheters [38]. The capsular material guards the bacterium from phagocytosis. Also it prevents the killing of the bacteria by bactericidal serum factors. [37] All the isolated Klebsiella strains showed resistance to cephalosporin antibiotics. This may be attributed to the expression of extended-spectrum β-lactamases (ESBLs) [39]. Klebsiella pneumoniae is a frequent cause of hospitalacquired and community-acquired urinary tract infections. The Klebsiella pneumoniae isolated from the hospital setting exhibited higher pathogenic potential, with higher genomic complexity. The hospital-acquired collection exhibited two specific virulence profiles VP13 (fimH, khe, mrkDV1) and VP22 (K2, fimH, khe, mrkDV1). High diversity and low accumulation of virulence genes in the same isolate was identified in Klebsiella pneumoniae community uropathogens. [40]. It has been narrated that multi drug-resistant Klebsiella pneumoniae is a usual cause of Urinary Tract Infections [41].
Although E. coli comprised only 15.7% of the isolates in the present study, in other reports, it was determined to be the primary pathogen in the urinary tract infection [42]. The interrelation between E. coli and UTI, though seemingly high, is decreasing slowly as E. coli is being substituted gradually by other members of the families Enterobacteriaceae and Enterococci [43].
Enterococci are habitually observed in the human gastrointestinal tract, mouth or vagina. The two most common enterococci in clinical samples are Enterococcus faecalis and Enterococcus faecium [44]. E. faecalis is one of the leading causes of human UTI and is the third most common hospital pathogen. It is responsible for ~12% of nosocomial infections and is a major etiological agent of CA-UTI. [45]. Pseudomonas aeruginosa belonging to the family Pseudomonodaceae was another predominant bacterial species in our study. Pseudomonas aeruginosa can be isolated from various living sources, including plants, animals, and humans. The ability of Pseudomonas aeruginosa to persist on minimal nutritional requirements and adapt to a range of physical conditions has endowed the organism to persist and cause both community-acquired infections and health care infections [46].
Morganella morganii belonging to the family Morganellaceae was also predominant in the study. It is commonly found in the environment and the intestinal tracts of humans, mammals, and reptiles as normal flora. It is an unusual opportunistic pathogen often encountered in the post-operative wound and urinary tract infections. Morganella morganii infections react satisfactorily to appropriate antibiotic therapy; yet, its natural resistance to many beta-lactam antibiotics may lead to the severity of such infections. Hence, Morganella morganii is a clinical pathogen that cannot be neglected due to its multiple antibiotic resistance [47].
Enterococcus of phylum Firmicutes is also prevalent along with the members of Enterobacteriaceae, Enterococci are habitually found in the human gastrointestinal tract, mouth or vagina. The two most common enterococci isolated in clinical samples are Enterococcus faecalis and Enterococcus faecium [44]. Enterococcus faecalis is the third most common hospital pathogen and one of the leading causes of human UTI [48], responsible for ~12% of nosocomial infections and is a significant etiological agent of CA-UTI. Enterococcus faecalis is also responsible for bacteremia, intra-abdominal infections and endocarditis. Enterococcal infections are difficult to treat due to the increasing incidence of multiple antibiotic resistance; Enterococci are intrinsically resistant to β-lactams and fluoroquinolones [45].
The antimicrobial sensitivity pattern confirmed that all of the urinary catheter isolates were resistant to the commonly used antibiotics including the second and third-generation cephalosporins and synthetic quinolones. Sensitivity to other commonly used antimicrobials including penicillin and fluoroquinolones were found to be lower than the semi-synthetic chloramphenicol and the aminoglycoside gentamycin. [49] EL-Banoby et al. reported that aminoglycosides were the most common antibiotics to which the organisms were sensitive particularly in the case of nosocomial UTI. Contrary to this report, Chloramphenicol was found to be the most effective antibiotic to the isolated clinical strains. Chloramphenicol is bacteriostatic for most Gram-positive and many Gram-negative aerobic bacteria. Its antibiotic activity is due to competitive inhibition for the binding of aminoacyl tRNA to the peptidyl transferase domain of the 50S subunit. Consequently, it induces a conformational change in the ribosome, which slows or even inhibits the incorporation of the aminoacyl tRNA and in turn the transpeptidation reaction. [50,51]. Gentamicin is a broad-spectrum antibiotic (except against streptococci and anaerobic bacteria) and it is bactericidal. It inhibits the bacterial protein synthesis by binding to 30S ribosomes. Gentamicin is indicated for acute serious infections caused by gramnegative bacteria [52].
The recurrent misuse of antibiotics in developing countries have resulted in changing antibiotic resistance profiles of bacterial populations [53]. The MAR index is a good risk assessment tool, with MAR index values > 0.2 indicating the high-risk source/site of contamination with antibiotic use [54]. All strong biofilm producers in this study showed a MAR index >0.2 implying an amplified incidence of MDR strains. The results showed that these organisms are likely to have originated from an environment where several antibiotics were used. Since the bacteria within biofilms are developing resistance to multiple ordinarily used antibiotics, treatment against CAUTI should also involve antimicrobial susceptibility tests of urinary catheter isolates [55].
Bacterial adhesion to xylene has been extensively used for measuring cell-surface hydrophobicity [56]. In this study, more than 60 per cent of the tested strains were strongly hydrophobic. As the urinary catheters are made from hydrophobic materials such as silicone, hydrophobic microorganisms can easily adhere to such surfaces. The effectiveness of biofilm formation is dependent on the cell surface hydrophobicity, as the hydrophobicity is important during the initial attachment process of bacteria to hydrophobic surfaces [25]. Aggregation shows a significant role in the formation of biofilms [57]. Autoaggregation generally protects the bacteria from external stresses such as nutrient starvation or oxidative stress [58]. In this study, the rate of auto aggregation increased with time in all the tested strains. Morganella morganii BTTDD6 showed high autoaggregation capabilities in comparison to the other strains. In this study, the strains Klebsiella pneumoniae BTTLA2, Klebsiella pneumoniae BTTDC5 ,Pseudomonas aeruginosa BTTDD3, Klebsiella pneumoniae BTTDD4 showed a positive connection between auto aggregation and cell surface hydrophobicity but there was no significant connection between auto aggregation and hydrophobicity in the case of Escherichia coli BTTDD5 and Morganella morganii BTTDD6.
Conclusion
For optimal antibiotic therapy of patients, identification and antibiotic susceptibility profiling of biofilm-associated bacteria in the patient catheter samples are inevitable. However, the multiple antibiotic resistance of biofilm-associated microbial pathogens makes the treatment difficult. The treatment options, specifically the choice of antibiotics can be made only after a careful evaluation of the antibiogram of the isolates. Another solution to fight the menace is to develop novel biofilm control strategies other than antibiotics such as bioactive compounds like bacteriocin, pyocyanin and phage therapy, to restrain the advent of antibiotic resistance.
Ethics and Informed Consent
Written informed consent was obtained from every patient for relevant experiments, and publication of the findings.
Acknowledgement
The authors wish to acknowledge Dr. Sunil. K. Narayanankutty, Department of Polymer Science and Rubber Technology, Cochin University of Science and Technology for proof reading the manuscript. This work was supported by the Council of Scientific and Industrial Research (CSIR), Government of India in the form of research fellowship and Department of Biotechnology, Cochin University of Science and Technology for the infrastructural support. The funding body played no role in the design of the study, collection, analysis, and interpretation of data or in writing the manuscript.
References
- Basak S, Rajurkar MN, Attal RO, Mallick SK. Biofilms: a challenge to medical fraternity in infection control. Infect Control. 2013.
- Thomsen TR, Hall-Stoodley L, Moser C, Stoodley P. The role of bacterial biofilms in infections of catheters and shunts. InBiofilm infections. 2011; 91- 109.
- Holá V, Ruzicka F, Horka M. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunology & Medical Microbiology. 2010; 59: 525-528.
- Donlan RM. Biofilms and device-associated infections. Emerging infectious diseases. 2001;7: 277.
- Stickler D, Morris N, Moreno MC, Sabbuba N. Studies on the formation of crystalline bacterial biofilms on urethral catheters. European Journal of Clinical Microbiology and Infectious Diseases. 1998; 17: 649-652.
- Rahman MM, Kim WS, Kumura H, Shimazaki KI. Autoaggregation and surface hydrophobicity of bifidobacteria. World Journal of Microbiology and Biotechnology. 2008; 24: 1593-1598.
- Hatt JK, Rather PN. Role of bacterial biofilms in urinary tract infections. Bacterial Biofilms. 2008: 163-192.
- Tenke P, Riedl CR, Jones GL, Williams GJ, Stickler D, Nagy E. Bacterial biofilm formation on urologic devices and heparin coating as preventive strategy. International journal of antimicrobial agents. 2004; 23: 67-74.
- Bryers JD. Medical biofilms. Biotechnology and bioengineering. 2008; 100: 1-8.
- Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cellular microbiology. 2009; 11: 1034-1043.
- Aslam S. Effect of antibacterials on biofilms. American journal of infection control. 2008; 36: S175-e9.
- Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Critical reviews in microbiology. 2009; 35: 340-355.
- Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. Journal of Clinical Pathology. 1989; 42: 872–874.
- Oliveira A, Maria de Lourdes RS. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC research notes. 2010; 3: 1-8.
- Rode TM, Langsrud S, Holck A, Møretrø T. Different patterns of biofilm formation in Staphylococcus aureus under food-related stress conditions. International journal of food microbiology. 2007; 116: 372-383.
- Shivaji S, Vijaya Bhanu N, Aggarwal RK. Identification of Yersinia pestis as the causative organism of plague in India as determined by 16S rDNA sequencing and RAPD-based genomic fingerprinting. FEMS microbiology letters. 2000; 189: 247-252.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990; 215: 403–410.
- Rzhetsky A, Nei M. A simple method for estimating and testing minimumevolution trees. 1992.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution. 2016; 33: 1870-1874.
- Bauer AT. Antibiotic susceptibility testing by a standardized single disc method. Am J clin pathol. 1966; 45: 149-58.
- Cockerill FR. Clinical, Institute LS. Performance standards for antimicrobial disk susceptibility testing: approved standard. National Committee for. Clinical Laboratory Standards. 2012.
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and environmental microbiology. 1983; 46: 165-170.
- Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92-Kos-2003-Journal of Applied Microbiology - Wiley Online Library n.d. 2003.
- Kwaszewska AK, Brewczynska A, Szewczyk EM. Hydrophobicity and biofilm formation of lipophilic skin corynebacteria. Polish journal of microbiology. 2006; 55: 189-193.
- Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs?. Frontiers in cellular and infection microbiology. 2014; 4: 112.
- Jain P, Parada JP, David A, Smith LG. Overuse of the indwelling urinary tract catheter in hospitalized medical patients. Archives of internal medicine. 1995; 155: 1425-1429.
- Saint S, Lipsky BA, Goold SD. Indwelling urinary catheters: a one-point restraint?. Annals of internal medicine. 2002; 137: 125-127.
- Tenney JH, Warren JW. Long-term catheter-associated bacteriuria: species at low concentration. Urology. 1987; 30: 444-446.
- Kaiser TD, Pereira EM, Dos Santos KR, Maciel EL, Schuenck RP, Nunes AP. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagnostic microbiology and infectious disease. 2013; 75: 235-239.
- Nicolle LE. Catheter-related urinary tract infection. Drugs & aging. 2005; 22: 627-639.
- Macleod SM, Stickler DJ. Species interactions in mixed-community crystalline biofilms on urinary catheters. Journal of medical microbiology. 2007; 56: 1549-1557.
- Cheryl-lynn YO, Beatson SA, Totsika M, Forestier C, McEwan AG, Schembri MA. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC microbiology. 2010; 10: 1-2.
- Wang X, Lünsdorf H, Ehrén I, Brauner A, Römling U. Characteristics of biofilms from urinary tract catheters and presence of biofilm-related components in Escherichia coli. Current microbiology. 2010; 60: 446-453.
- Jarvis WR, Munn VP, Highsmith AK, Culver DH, Hughes JM. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infection Control & Hospital Epidemiology. 1985; 6: 68-74.
- Spencer RC. Predominant pathogens found in the European prevalence of infection in intensive care study. European Journal of Clinical Microbiology and Infectious Diseases. 1996; 15: 281-285.
- Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical microbiology reviews. 1998; 11: 589-603.
- Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014; 3: 743-758.
- Boddicker JD, Anderson RA, Jagnow J, Clegg S. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infection and immunity. 2006; 74: 4590-4597.
- Grover SS, Sharma M, Chattopadhya D, Kapoor H, Pasha ST, Singh G. Phenotypic and genotypic detection of ESBL mediated cephalosporin resistance in Klebsiella pneumoniae: emergence of high resistance against cefepime, the fourth generation cephalosporin. Journal of Infection. 2006; 53: 279-288.
- Caneiras C, Lito L, Melo-Cristino J, Duarte A. Community-and hospitalacquired Klebsiella pneumoniae urinary tract infections in Portugal: virulence and antibiotic resistance. Microorganisms. 2019; 7: 138.
- Sherif M, Palmieri M, Mirande C, El-Mahallawy H, Rashed HG, Abd-El- Reheem F, et al. Whole-genome sequencing of Egyptian multidrug-resistant Klebsiella pneumoniae isolates: a multi-center pilot study. European Journal of Clinical Microbiology & Infectious Diseases. 2021: 1-0.
- Alanazi MQ, Alqahtani FY, Aleanizy FS. An evaluation of E. coli in urinary tract infection in emergency department at KAMC in Riyadh, Saudi Arabia: retrospective study. Annals of clinical microbiology and antimicrobials. 2018; 17: 1-7.
- Teshager L, Asrat D, Gebre-Selassie S, Tamiru S. Catheterized and noncatheterized urinary tract infections among patients attended at Jimma University Teaching Hospital, Southwest, Ethiopia. Ethiopian medical journal. 2008; 46: 55-62.
- Bello Gonzalez TD, Pham P, Top J, Willems RJ, van Schaik W, van Passel MW, et al. Characterization of Enterococcus isolates colonizing the intestinal tract of intensive care unit patients receiving selective digestive decontamination. Frontiers in microbiology. 2017; 8: 1596.
- Marra A, Dib-Hajj F, Lamb L, Kaczmarek F, Shang W, Beckius G, et al. Enterococcal virulence determinants may be involved in resistance to clinical therapy. Diagnostic microbiology and infectious disease. 2007; 58: 59-65.
- Kaier K, Heister T, Götting T, Wolkewitz M, Mutters NT. Measuring the inhospital costs of Pseudomonas aeruginosa pneumonia: methodology and results from a German teaching hospital. BMC Infectious Diseases 2019; 19: 1028.
- Liu H, Zhu J, Hu Q, Rao X. Morganella morganii, a non-negligent opportunistic pathogen. International Journal of Infectious Diseases. 2016; 50: 10-17.
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature reviews microbiology. 2015; 13: 269-284.
- El-Banoby MH, Ebeid SM, El-Bedewy RS, Amrousy MA. Nosocomial pneumonia and urinary tract infections in elderly patients admitted to the ICU. Egypt J Med Lab Sci. 2007; 16: 51–56.
- Milosevic TV, Payen VL, Sonveaux P, Muccioli GG, Tulkens PM, Van Bambeke F. Mitochondrial alterations (inhibition of mitochondrial protein expression, oxidative metabolism, and ultrastructure) induced by linezolid and tedizolid at clinically relevant concentrations in cultured human HL-60 promyelocytes and THP-1 monocytes. Antimicrobial agents and chemotherapy. 2018; 62.
- Maddison JE, Page SW, Church DB, editors. Small animal clinical pharmacology. Elsevier Health Sciences. 2008.
- Fitzgerald KT, Newquist KL. Poisonings in the captive reptile. Small Animal Toxicology. 2013: 229-249.
- Byarugaba DK. Antimicrobial resistance in developing countries and responsible risk factors. International journal of antimicrobial agents. 2004; 24: 105-110.
- Osundiya OO, Oladele RO, Oduyebo OO. Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African Journal of Clinical and Experimental Microbiology. 2013; 14: 164-168.
- Osungunna MO, Onawunmi GO. Antibiotic resistance profiles of biofilmforming bacteria associated with urine and urinary catheters in a tertiary hospital in Ile-Ife, Nigeria. Southern African Journal of Infectious Diseases. 2018; 33: 80-85.
- Lajhar SA, Brownlie J, Barlow R. Characterization of biofilm-forming capacity and resistance to sanitizers of a range of E. coli O26 pathotypes from clinical cases and cattle in Australia. BMC microbiology. 2018; 18: 1-5.
- Tuo Y, Yu H, Ai L, Wu Z, Guo B, Chen W. Aggregation and adhesion properties of 22 Lactobacillus strains. Journal of dairy science. 2013;96: 4252-4257.
- Trunk T, Khalil HS, Leo JC. Bacterial autoaggregation. AIMS microbiology. 2018; 4: 140-164.