Research Article
Austin J Microbiol. 2021; 6(1): 1031.
Antimicrobial Resistant Salmonella Serotypes Circulating in Meat in Senegal
Sow O¹, Gbe KM², Ndiaye I¹, Cissé A¹, Sy AC², Wane AA¹, Seck A³ and Sambe B¹*
¹Experimental Bacteriology Unit, Pasteur Institute Dakar, Senegal
²Laboratory of Food Safety and Environmental Hygiene, Pasteur Institute of Dakar, Senegal
³Laboratory of Medical Biology, Pasteur Institute Dakar, Senegal
*Corresponding author: Bissoume Sambe, Experimental Bacteriology Unit, Pasteur Institute of Dakar, 36, Avenue Pasteur, Dakar, Zip: 220, Senegal
Received: May 25, 2021; Accepted: June 19, 2021; Published: June 26, 2021
Abstract
The Presence of antimicrobial-resistant Salmonella in food remains a real threat. The aim of this study was to determine the antimicrobial resistance profiles of Salmonella strains isolated from the most consumed types of meat in Senegal.
A total of 337 samples were collected including 247 samples of raw meat and 90 of meat-based ready-made meals. Salmonella strains were isolated according to the French standard EN-ISO-6579 and serotyped according to the KWLM scheme. Antimicrobial susceptibility tests were performed with 16 discs following the agar diffusion method.
Our study showed that 37% of the samples carried Salmonella and allowed the isolation of 136 strains of Salmonella. We identified 47 different serotypes including S. Kentucky (11/136: 8%), S. Brancaster (10/136: 7%), S. Chester (10/136: 7%), S. Istanbul (5/136: 4 %), and S. Agona (5/136: 4%) which were respectively the most common. Of the 136 strains, 43 (31.6%) were resistant to at least one antibiotic, of which 15.4% (21/136) were multidrug-resistant. Interestingly, S. Kentucky that was the most common, exhibited also the highest level of resistance, being at least resistant to 8 antibiotics.
In conclusion, The increasing resistance and spread of S. Kentucky in meat products demand special monitoring and to be further studied.
Keywords: Meat; chicken broilers; Salmonella serotypes; Antimicrobialresistance; Senegal
Abbreviations
BPW: Buffered Peptone Water; S.: Salmonella; KWLM: Kauffmann-White-Le-Minor; MKTTn: Mueller Kauffmann Tetrathionate Novobiocin Broth; XLD: Xylose Lysine Deoxycholate
Introduction
Salmonella are particularly pathogenic microorganisms that infect both animals and humans; they can be transmitted between animals and humans through direct contact or contaminated-food [1,2]. They are often carried by water drink and vegetables but food of animal origins are the major vehicles [3-5]. They are responsible, after oral absorption, for typhoid and paratyphoid fevers, and gastroenteritis [6,7]. Although the typhoid and paratyphoid fevers are more severe, they are getting rare in some regions of the world [8]. On the other hand, Non Typhoidal Salmonella (NTS) are of big clinical concern due to the huge number of people they infect annually and the cost of medical care [1,9]. Moreover, in certain cases, NTS can spread up to other tissues of the body and cause severe illness [1]. This may turn in to fatal infection for people with weak immune system (infants, patients with immunodeficient or immunosuppressed systems, etc.). In fact, foodborne salmonellosis has been estimated at more than 80 million cases and is believed to be associated with thousands of deaths each year worldwide, thus constituting a major public health problem [9,10].
The threat has become much more serious with the spread of multidrug resistant Salmonella strains in food. Several studies have shown evidence that meat is potential source of salmonellosis when adequate hygiene and sanitation are lacking in the food management [4,11]. Yet, due to changing climatic conditions and sedentary lifestyles, people are feeding mainly on animal products. According to FAO [12], world meat production is expected to be 16% higher in 2025 than the reference period, 2013-2015. Therefore, this increase in the consumption of meat requires surveillance of antimicrobialresistant Salmonella in this sector knowing that they are the bacterial leading cause of foodborne disease [11].
In 2014, WHO had classified antimicrobial resistance as one of the three most threatening health problems of the 21st century [13]. This problem of AMR requires the establishment of surveillance systems to monitor its evolution. The epidemiological data collected would make it possible to estimate the antimicrobial resistance patterns of Salmonella strains and to alert the authorities to the implementation of anticipatory policies. In this context, we aimed to study foodborne Salmonella isolated from raw meat and meat-based ready-made meals in Senegal. To our knowledge, this is the first study tackling the problem of the antimicrobial-resistant Salmonella strains carried in the three main types of meat consumed in Senegal.
The general objective of this study was to provide epidemiological data on the antimicrobial resistance of Salmonella strains isolated from the 3 main types of meat consumed in Senegal. The specific objectives were (i) to assess the level of Salmonella carriage of chicken broilers, beef-, sheep-meats, and meat-based ready-made meals, (ii) to identify the different serotypes and (iii) to determine the antimicrobial resistance profiles of the isolates.
Materials and Methods
Sampling
The samples were randomized and collected from different slaughterhouses and markets in the urban and peri-urban zone of Dakar. During the period January 2015 to May 2016, a total of 337 samples were collected including 247 samples of raw meat (Chicken broiler: 60, beef meat: 100 and sheep meat: 87) and 90 samples of meatbased ready-made meals. The samples were immediately transported into iceboxes to the Laboratory of Hygiene and Environment Safety of Pasteur Institute Dakar. All samples were stored at -80°C after they were analysed.
Salmonella screening
According to the French standard EN ISO 6579 for Salmonella assessment in food, 25 g of each sample was mixed with a STOMACHERND mill in 225 mL of Buffered Peptone Water (BPW) and then the mixture was incubated at 37°C for 18 hours. From these pre-enrichment (BPW culture), 0.1 mL was transplanted into a tube containing 10ml of Rappaport Vasiliadis broth (RVS) and 1 mL (BPW culture) into 9 mL of Mueller Kauffmann Tetrathionate Novobiocin Broth (MKTTn). Tubes containing RVS broth were incubated at 41.5°C in a stirred bath and those of MKTTn in an oven at 37°C for 24 hours. The culture of each broth was parallelly isolated on two different selective-agar-media, Xylose Lysine Deoxycholate (XLD) and Hektoen and then incubated at 37°C. After 24 hours of incubation, colonies with a black center with halo on Hektoen and a black center surrounded by a reddish zone, slightly transparent on XLD were considered as Salmonella presumptive-colonies. Thereby, five (5) presumptive colonies were isolated on Nutrient agar then incubated at 37°C for 18 hours. The oxidase test was done with strip kits before using the API 20E gallery intended to the biochemical identification of Enterobacteriaceae (Biomérieux). As Salmonella are oxidase negative, only strains with negative oxidase test, were considered. From the Nutrient agar with negative oxidase test, a pure colony has been suspended in 5 mL of physiological saline solution and distributed in the different wells of the gallery API 20E. The gallery has been incubated at 37°C for 24 h and read with apiweb software according to the protocol API 20E (Biomérieux). The identified strains were stored at -80°C in TCS + Glycerol 10%.
Serotyping
The serotyping was done according to the White Kauffmann Le Minor scheme (WKLM) 2007. So, Salmonella strains were cultured at a while on Mueller Hinton [14] and Kliger Hajna (KH) agars for the determination of the somatic and flagellar phases, respectively. KH medium (Bio-Rad) in which 1mL of physiological saline solution was added, was used to promote flagellar development.
Determination of the somatic phase: The (pure) MH culture was used to do the agglutination tests with the polyvalent antisera (OMA, OMB, OMC, OMD and OME) to determine the group to whom the isolate belong. Whenever an isolate had agglutinated with one of the polyvalent, it was confronted with the different corresponding monovalent-antisera that make up this group to determine its somatic phase.
Determination of flagellar phases: As in most cases, Salmonella have two (2) flagellar phases. The first to appear was noted phase H1 and the second, H2. In rare cases, there is a third phase. KH Culture was used to determine the flagellar phases. Each strain of Salmonella was tested against 5 polyvalent antisera HMA, HMB, HMC, HMD and HME. The strains having agglutinated with two different polyvalent, were confronted with the monovalent antisera that compose respectively these two polyvalent antisera to determine directly the phases H1 and H2. For a strain not simultaneously expressing its two phases, its H1 phase was first determined, then a phase inversion was performed to determine the second phase H2 (Figure 1). Sven Guard soft agar (Bio-Rad), poured into 55 cm Petri dishes. This soft agar was used to demonstrate the inapparent H2 antigen phase of biphasic Salmonella. A drop of the monovalent which was used for the determination of H1 was deposited in the center of the soft agar Sven Gard then a drop of culture of the same strain was added on and then, the culture was incubated at 37°C/24h. This aimed to block the H1 phase to allow the development of the H2 phase. During incubation, the strain developed its H2 phase to migrate. 24 hours later, the strain that had developed its H2 phase had migrated to the edges of the plate (Figure 1A). The determination of the H2 phase was therefore made by making the agglutination tests from the bacterial growth at the periphery of the box.
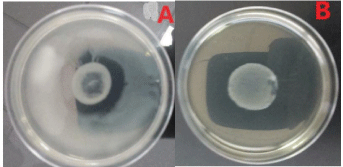
Figure 1: Flagellar phases of Salmonella.
A. Salmonella Kentucky: Trapping of the first phase (dark white in the center)
and development of the second (whitish layer at the periphery).
B. Salmonella Gueuletapee: Trapping of the first phase (dark white in the
center)-An absence of the second phase.
After determining the somatic and flagellar phases of the strain, its antigenic formula was written as follows: O: H1; H2. This formula was used to determine the serovar to which it corresponds, referring to the WKLM Scheme of 2007 (example: 38: eh, 1,2 corresponds to Salmonella Thiaroye). Some strains had developed a single flagellar phase and were noted as O: H1; - (Example 4: fgs; - corresponds to Salmonella Agona) (Figure 1B). Others had three phases (O: H1, H2, H3 Example 8.20: r [i], z6: Salmonella Altona). The strains that this method failed to serotype, will be sent to the National Reference Center of the Pasteur Institute of Paris for a further characterization
Antimicrobial susceptibility tests
The antimicrobial susceptibility was determined by agar diffusion method using the Mueller Hinton agar (Bio-Rad). Sixteen (16) antibiotics disks (Bio-Rad) were chosen from the standard list proposed by EUCAST version 2017, based on their importance both in human and veterinary medicine. The position of the amoxicillin+clavulanic acid regarding the third and fourth cephalosporins’ generations, aimed at detecting an eventual ESBL (Extended-Spectrum Beta-lactamases). Antibiotics susceptibility were read with the automated system Adagio (Bio-Rad). The isolates were categorized “susceptible”, “intermediate” or “resistant” based on their inhibitory diameters referring to EUCAST 2017 standards. Parallelly, a check was made at the same time with a reference strain to validate the tests.
Data management and exploitation
Sampling data and experimental results were collected and managed with Excel software (Office 2016). The R software was used to perform the statistical tests but some graphical representations were made with Excel (Office 2016).
Results and Discussion
Contamination rates and serovars distribution
Bacteriological analysis of different samples showed that 36.79 (124/337) of samples were contaminated with Salmonella (Table 1). Chicken broilers exhibited the highest rates of Salmonella carriage (58%) followed by the bovine and ovine meat (~41%). Out of the 90 sample of meat-based ready-to-use meals, 13% was contaminated with Salmonella (Table I).
Sample Type
Assessed Samples
Positive Samples
Chicken broilers
60
35
Ovine meat
87
37
Bovine meat
100
40
Ready-to-use meals
90
12
Total
337
124
Table 1: Level of meat contamination with salmonella.
A total of 136 Salmonella spp were recovered out of which 126 were completely serotyped. Hence, 47 different serovars were identified of which Salmonella Kentucky (11/136: 8%), Salmonella Brancaster (10/136: 7%), Salmonella Chester (10/136: 7%), Salmonella Istanbul (5/136: 4%) and Salmonella Agona (5/136: 4%), were the most common serovars (Supplementary, Table 1). However various local serovars (Salmonella Ngor, Salmonella Fann, Salmonella Gueuletapee, Salmonella Thiaroye and Salmonella Gambia) have been found in beef. Three (03) serovars of Salmonella enterica subsp. salamae and two (02) of Salmonella enterica subsp. arizonae were also found in beef.
Antimicrobial resistance profiles
Strains isolated from sheep-meat, beef-meat and cooked dishes were almost susceptible to all antibiotics tested: 33/34; 36/42 and 18/20 respectively. On the other hand, 34 out of the 40 strains isolated from broilers, were resistant and then represented 80% of the total resistant strains (Figure 2).
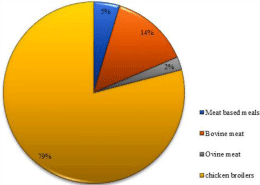
Figure 2: Distribution of the resistant-strains based on the food origin.
All the strains were susceptible to chloramphenicol and to imipenem. No ESBL has been observed. However, resistance to several first-line antibiotics has been observed. In fact, of the 136 strains, 43 (31.62%) were resistant to at least one antibiotic. Out of these 43 strains, 48.8% (21/43) presented resistance to at least 4 antibiotics. Two (02) serovars of Salmonella Kentucky showed the highest level of resistance being resistant to at least 8 antibiotics (Table 2).
Serovars
Number
Resistance profiles
Brancaster
6
TETR
Brancaster
2
TETR, SXTR
Konstanz
1
TETR, SXTR
Djugu
1
TETR, SXTR
Brandenburg
1
TETR, SXTR
Chester
6
TETR, SXTR
Bredeney
1
AMPR, TICR, AMCR
Unknown
3
AMPR, AMCR, CEFR, FOXR
Chester
4
TETR, SXTR, NAR, NORR, CIPR
Johannesburg
3
TETR, SXTR, NAR, NORR, CIPR
Istanbul
5
TETR, SXTR, NAR, NORR, CIPR
Kentucky
3
TETR, SXTR, GMR
Godeau
1
TETR, SXTR, AMPR, TICR AMCR
Myrria
1
TETR, SXTR, AMPR, TICR AMCR
Salford
1
TETR, SXTR, AMPR, TICR AMCR
Kentucky
2
TETR, SXTR, GMR, NAR, NORR, CIPR, AMPR, TICR et AMCR
R= Resistant to (E.g. TETR = resistant to TET); TET=Tetracyclin; SXT=Trimethoprim+Sulfamethoxazole; AMP= Ampicillin; TIC=Ticarcillin; AMC =Amoxillin+Clavulanic acid; CEF=Cefalotin; FOX=Cefoxitin; NA=Nalidixic acid; NOR=Norfloxacin; CIP=Ciprofloxacin; GM= Resistant to Gentamicin.
Table 2: Resistance levels based on serovars types.
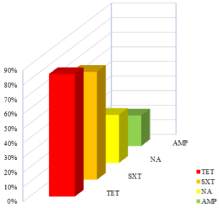
High frequency of resistance to tetracycline and trimethoprimsulfamethoxazole, being 83.72% (36/43) and 74.42% (32/43) respectively, has been noted. Resistance to fluoro/quinolones (32.56%), and to ampicillin (20.93%) was also relatively frequent (Figure 3).

Figure 3: Most significant resistance patterns.
AMP=ampicillin; NA=Nalidixic acid; SXT=trimethoprim+Sulfamethoxazole;
TET=tetracyclin.
Discussion
Our results have shown that in Senegal although cooked dishes have a high level of contamination with Salmonella (13%), meat from bovine and ovine origins are much more contaminated (40 to 42%). But the most alarming contamination rates were noted with broilers (58%). The skins of livestock animals are often a source of contamination for meat when strict hygiene measures are not taken at slaughter. They carry Salmonella from environmental, faecal and other sources. To these external contaminants must be added the internal microbial flora of these animals which can also contaminate the meat during slaughter and evisceration. This would justify Salmonella contamination rates we are reporting here. Our study corroborates that of another study conducted in Senegal in 2017 which showed that these two types of meat are at 40% contaminated with Salmonella (Unpublished data). Such levels of contamination have been reported before. According to this study, the contamination rates of beef and sheep meat were 56.6% and 66.6% respectively (Unpublished data).
The conditions for rearing, processing and slaughtering cattle and sheep are almost identical in Senegal, which may be the reason why their levels of contamination have the same tendency. These conditions differ from those of broilers. Indeed, the breeding conditions of chickens (damp and nutrient-rich hen houses) are a primary source of Salmonella contamination for the broilers. Then come slaughter and plucking, which are critical steps for the microbiological quality of the chickens. Indeed, they are frequently the cause of an increase in the bacterial load of the chicken due to unhygienic handling and cross contamination. This same situation would reoccur at the market where chickens are piled up and exposed for sale. This would explain the high level of contamination that we have found (58%). Other studies conducted in Senegal, reported similar results. According to, Fall-Niang and colleagues, 53% of the broilers were contaminated with Salmonella. The study of Diouf also revealed as high contamination rates (52%) [15]. Similar results have been found elsewhere in the world: 60% in Cameroon [16] and 54% in Japan [17]. Although the meat products used in the preparation of shawarma, sandwiches and other dishes are contaminated in the same way as raw meats (Table III), the cooking heat considerably reduces these contaminants. Salmonella is almost eliminated at 70°C, so ready meals are less contaminated than raw meat. Moreover, these contaminations may come mainly from a post-cooking source. This is why we would obtain a relatively low but not negligible contamination rate (13%). Such results were found by El Marnissi in Morocco, accounting for 13.70% of cooked dishes [18].
From the 124 contaminated samples (124/337), 136 Salmonella strains were isolated. In fact, in some samples, 2 to 3 different Salmonella have been isolated which would explain the fact that there are more strains than contaminated samples.
Among the 136 strains isolated, 47 different serovars were identified of which Salmonella Kentucky, Salmonella Chester, and Salmonella Brancaster were the most common serovars, respectively. Our results are similar to those of some authors in Africa. Alambedji and colleagues had previously shown that S. Kentucky, S. Muenster and S. Brancaster were the most common in chicken carcasses in Dakar [19]. A recent study conducted in Senegal have also found that the serotypes Istambul, Brancaster and Kentucky were respectively the most prevalent in the broilers [20]. The difference found in the frequency of the serovars by these authors compared to our study could be explained by the fact the they only studied strains of chicken origin. Another study in Nigeria showed the same trend [21]. However, 7% of our strains are not completely typified, therefore, this some serovars dominance over others could be switched.
The selection pressure and the adaptation need of the strains have led to the emergence of resistant clones that propagate this resistance. Our study hav showed a relatively high level of resistance (31.62%). Indeed, our results are similar to those of Bouchrif and Ejo.
The study of Bouchrif et al. on Salmonella strains isolated from chickens broiler, beef and cooked dishes in Morocco showed similar levels of resistance ,being 29% [22]. In addition, a recent study in Ethiopia showed a high prevalence of resistant Salmonella [23]. This prevalence is much higher than our results. However, this difference (72.7% versus 31.62%) could be justified by the fact that their study only concerned Salmonella isolated from broilers where the prevalence of antimicrobial resistance is generally very high. Similar results would be obtained if we considered separately our strains isolated from chickens because 85% of these isolates are at least resistant to one antibiotic. In fact, according to a study conducted in Dakar in 2006, about 79% of Salmonella isolated from chickens were resistant to at least one antibiotic [19].
It appears that the level of sensitivity of the strains would also depend on its immediate environment, the matrix from which they were isolated. Strains that have evolved in different environments would have acquired different mechanisms of resistance, materialized by mutations (gain or loss of genetic determinants) within their genomes. This could result in a difference in antibiotic susceptibility of strains isolated from animals exposed to antibiotics compared to those isolated from unexposed or less exposed animals.
The results we obtained show that the resistance profiles of strains isolated from broilers are significantly different from those of isolates from other matrices (p-value<0.0001). In Senegal, sheep and cattle have very little or no exposure to antibiotics, which explains the poor evolution of strains isolated from these sectors from the point of view of antibiotic resistance. The cooked dishes we have studied show the same profiles since they are mainly made from beef.
The level of resistance of the isolates therefore depends not only on the bacterial species but also on several other factors. It is clear that exposure to antibiotics is accelerating the phenomenon of antimicrobial resistance [24]. In the poultry sector, for example, farmers are massively using antibiotics as both treatments and feed for poultry. This may select the bacteria that acquired resistance to these antibiotics. This new resistant and / or multidrug-resistant population is developing and becoming predominant in these areas. This explains the high levels of resistance obtained in our work from broilers (85%). These results are consistent with those of several other studies done in Senegal (Bada-Alambedji et al., 2006) and in the rest of the world [25-27]. In Brazil, a study of chicken carcasses showed a higher resistance rate with 100% resistance [28]. This may be due to the fact that Brazil is one of the five countries with the highest antibiotic consumption in the livestock sector, alongside China, the United States, Germany and India [29]. However, in Senegal there has been a marked increase in the use of antibiotics in breeding in recent years. The amount of antibiotics used in livestock farming in Senegal increased from 11,435 kg in 2015 to 14,547 kg in 2017, an increase of nearly 3,112 tones [30].
Resistance to 3rd and 4th generation cephalosporins or carbapenems was not observed. Such results have been found here in Senegal [31-33], in Africa [22,23], and elsewhere [34]. However, ESBL-producing Salmonella in food, has been previously described in Senegal [35]. Another interesting finding of our study is that all the strains was susceptible to chloramphenicol giving more choices to the clinicians. This is consistent with previous studies carried in Senegal [20,36].
Unlike these antibiotics, resistance to penicillin (AMP, TIC), tetracyclines [14] and sulfonamides, are widely distributed within Salmonella strains. Added to this is the resistance to fluoro/quinolones more and more found in these strains. Significant resistance profiles for tetracycline (83.72%), trimethoprim-sulfamethoxazole (74.42%), quinolones/fluoroquinolones (32.56%), and ampicillin (20.93%) were found. These trends are reported in the literature by many authors [23,37,38].
Conclusion
This work allowed us to estimate the antimicrobial resistance level of the Salmonella strains isolated from the most consumed meat in Dakar. One most the most important findinds is that Salmonella Kentucky that is the most prevalent serovar, exhibits the highest level of resistance being at least resistant to eight (08) antibiotics. These data would make it possible to prevent “therapeutic failures” during a possible salmonellosis of meat origin in Senegal. They would also alert the public and the competent authorities and lay the groundwork for the foundation of a plan to monitor this resistance.
Ethical Statements
This study doesn’t involve any human or animal testing.
Authors Contributions
NI, CA and SAC helped to collect samples and participated to some lab works. WAA contributed to set and control the bacterial analyses. GKM was involved in strains isolation and helped to perform the Salmonella serotyping. SA contributed to antimicrobial resistance profiles interpretation. SBB supervised the results analysis and interpretation and revised the manuscript. SO is the main author hence, participated to the experiment design, performed the lab work, did the bibliographic review, analysed and interpreted the results, designed the figure and drafted the maniscrit.
All the authors have read and approved the submitted version of the maniscrit.
Acknowlegement
The authors are grateful to late Professor Amy Gassama-Sow and address particular acknowledgments to her. She designed and supervised the whole project. She was the Head of the Experimental bacteriology Unit and Laboratory of Food Safety & Environmental Hygiene. She passed away in June 2019. May his Soul rest in perfect Peace!
Lot of thanks to Dr. Nawmin Siourimè Somda for reviewing the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Sterzenbach T, Crawford RW, Winter SE, Baümler AJ. Salmonella virulence mechanisms and their genetic basis, in Salmonella in domestic animals. 2013; 80.
- Rabsch W, Simon S, Humphrey T. Public health aspects of Salmonella infections, in Salmonella in domestic animals. 2013; 351-376.
- Barrow PA, et al. The long view: Salmonella – the last forty years. Avian Pathology. 2012; 41: 413-420.
- Bardon J, Ondruskova J, Ambroz P. [Prevalence of Salmonella in meat and meat products in Moravia in 2010-2015]. Klin Mikrobiol Infekc Lek, 2016. 22: 48-53.
- Budd W. Typhoid fever its nature, mode of spreading, and prevention. Am J Public Health (NY). 1918; 8: 610-612.
- Crump JA. Progress in Typhoid Fever Epidemiology. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2019; 68: S4-S9.
- McWhorter AR, Davos D, Chousalkar KK. Pathogenicity of Salmonella strains isolated from egg shells and the layer farm environment in australia. Applied and environmental microbiology. 2015; 81: 405-414.
- Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: Systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012; 2: 010401.
- Majowicz SE, et al. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin Infec Dis. 2010; 50: 882-889.
- Wang H, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet. 2016; 388: 1459-1544.
- Feasey NA, et al. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet (London, England). 2012; 379: 2489-2499.
- OECD/FAO. Perspectives Agricoles de l’OCDE et de la FAO 2016-2025. Food and Agriculture organization of the United Nations. 2016.
- WHO, Antimicrobial resistance: global report on surveillance 2014. 2014.
- Wang H, et al. Global, regional, and national life expectancy, allcause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet. 2016; 388: 1459-1544.
- Diouf KCN. Surveillance de la résistance aux antibiotiques des souches de salmonella spp et Escherichia coli isolées de la viande de poulet de chair au Sénégal. Université Cheikh Anta Diop de Dakar. 2006.
- Wouafo M, et al. Prevalence and antimicrobial resistance of Salmonella serotypes in chickens from retail markets in Yaounde (Cameroon). Microb Drug Resist. 2010; 16: 171-176.
- Furukawa I, et al. Prevalence and Characteristics of Salmonella and Campylobacter in Retail Poultry Meat in Japan. J Infect Dis. 2017; 70: 239- 247.
- El Marnissi B, et al. Contribution a l’etude de la qualite microbiologique de denrees alimentaires commercialisees a fes-boulemane. 2012; 6: 98-117.
- Bada-Alambedji R, et al. Antimicrobial resistance of Salmonella isolated from poultry carcasses in Dakar (Senegal). Brazilian Journal of Microbiology. 2006; 37: 510-515.
- Fall-Niang NK, et al. Antimicrobial Resistance Profile of Salmonella Isolates in Chicken Carcasses in Dakar, Senegal. Foodborne Pathog Dis. 2019; 16: 130-136.
- Fagbamila IO, et al. Salmonella serovars and their distribution in Nigerian commercial chicken layer farms. PLoS ONE. 2017; 12: e0173097.
- Bouchrif B, et al. Prevalence and antibiotic-resistance of Salmonella isolated from food in Morocco. J Infect Dev Ctries. 2009; 3: 35-40.
- Kassahun Tessema HB, Mebrat Ejo, Adem Hiko. Prevalence and Antibiotic Resistance of Salmonella Species Isolated from Chicken Eggs by Standard Bacteriological Method. Journal of Veterinary Science & Technology. 2017; 8: 1-5.
- CDDEP. Antibiotic use. Resistance Map. 2015.
- Pulido-Landinez M, et al. Serotype and antimicrobial resistance patterns of Salmonella isolates from commercial birds and poultry environment in Mississippi. Avian Dis. 2014; 58: 64-70.
- Yang B, et al. Counts, serotypes, and antimicrobial resistance of Salmonella isolates on retail raw poultry in the People’s Republic of China. J Food Prot. 2014; 77: 894-902.
- Mattiello SP, et al. Characterization of antimicrobial resistance in Salmonella enterica strains isolated from Brazilian poultry production. Antonie Van Leeuwenhoek. 2015; 108: 1227-1238.
- Medeiros MA, et al. Prevalence and antimicrobial resistance of Salmonella in chicken carcasses at retail in 15 Brazilian cities. Rev Panam Salud Publica. 2011; 30: 555-560.
- Van Boeckel TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015; 112: 5649-5654.
- FAO. La FAO invite les vétérinaires et para-vétérinaires à une utilisation responsable des antimicrobiens. 2018.
- Dione MM, et al. Prevalence and antimicrobial resistance of Salmonella isolated from broiler farms, chicken carcasses, and street-vended restaurants in Casamance, Senegal. J Food Prot. 2009; 72: 2423-2427.
- Dione MM, Geerts S, Antonio M. Characterisation of novel strains of multiply antibiotic-resistant Salmonella recovered from poultry in Southern Senegal. J Infect Dev Ctries. 2012; 6: 436-442.
- Cardinale E, et al. Epidemiological analysis of Salmonella enterica ssp. enterica serovars Hadar, Brancaster and Enteritidis from humans and broiler chickens in Senegal using pulsed-field gel electrophoresis and antibiotic susceptibility. J Appl Microbiol. 2005; 99: 968-977.
- Sodagari HR, Mashak Z, Ghadimianazar A. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from retail chicken meat and giblets in Iran. J Infect Dev Ctries. 2015; 9: 463-469.
- Gassama-Sow A, et al. Integrons in Salmonella Keurmassar, Senegal. Emerging Infectious Diseases. 2004; 10: 1339-1341.
- Bada-Alambedji R, et al. Antimicrobial resistance of Salmonella isolated from poultry carcasses in Dakar (Senegal). 2006; 37: 510-515.
- Diouf KCN. Surveillance de la resistance aux antibiotiques des souches de Salmonella spp et Escherichia coli isolees de la viande de poulet de chair au senegal in Ecole Inter-états des Sciences et Medecine Vétérinaires. Université Cheikh Anta Diop de Dakar. 2006.
- Yu T, et al. Antimicrobial resistance, class 1 integrons, and horizontal transfer in Salmonella isolated from retail food in Henan, China. J Infect Dev Ctries. 2014; 8: 705-711.