
Research Article
Austin J Microbiol. 2022; 7(1): 1040.
Molecular Characterization of Extended-Spectrum Beta-Lactamase-Producing Extra-Intestinal Pathogenic Escherichia coli Isolated in a University Teaching Hospital Dakar-Senegal
Dossouvi KM1*, Sambe-Ba B2, Lo G1,3, Cissé A2, Ba-Diallo A1,3, Ndiaye I2, Dieng A1, Ndiaye SML1, Fall C2, Tine A1, Karam F1, Diagne-Samb H1, Ngom-Cisse S1, Diop-Ndiaye H1,3, Toure-Kane C3, Gaye-Diallo A1,3, Mboup S1,3, Boye CSB1, Dièye Y2, Seck A2,4 and Camara M1,3
1Bacteriology-Virology Laboratory, National University Hospital, Aristide Le Dantec, Dakar, Senegal
2Pole of Microbiology, Institut Pasteur de Dakar, Senegal
3Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formation (IRESSEF), Dakar, Senegal
4Medical Analysis Laboratory, Institut Pasteur de Dakar, Senegal
*Corresponding author: Dossouvi KM, Bacteriology-Virology Laboratory, National University Hospital, Aristide Le Dantec– 30, avenue Pasteur-BP7325-Dakar, Sénégal
Received: July 29, 2022; Accepted: August 25, 2022; Published: September 01, 2022
Abstract
Extra-Intestinal Pathogenic Escherichia coli (ExPEC) is a predominant Gram-negative bacterial pathogen and is responsible of several diseases including Urinary Tract Infections (UTI), nosocomial pneumonia, and neonatal meningitis. ExPEC isolates are often multidrug resistant and clones producing Extended-Spectrum Beta-Lactamases (ESBL) are increasingly reported all over the world.
Seventy-eight clinical ExPEC strains (49 Community-Acquired (CA) and 29 Hospital-Acquired (HA)) were selected for this study. The majority was from UTIs (n=51), while the rest (n=27) was from pus, sputum, bronchial fluid and vaginal samples (non-uropathogenic ExPEC). Antibiotic susceptibility testing was performed using the Kirby-Bauer disc diffusion method. Standard polymerase chain reaction was used to screen major ESBL genes (blaCTX-M, blaOXA-1, blaTEM, blaSHV) and blaCTX-M variants (blaCTX-M-1, blaCTX-M-9, blaCTX-M-15, blaCTX-M-25).
All the tested isolates were resistant to ampicillin, ticarcillin, amoxicillin/ clavulanic acid combination, cefalotin, cefotaxime, ceftazidime, cefepime and aztreonam, but showed a high susceptibityto fosfomycin (98.7%, n = 77), ertapenem (96.2%, n = 75), and imipenem (100%). Moreover, isolates harbored at least one ESBL gene, including blaCTX-M (98.7%), blaOXA-1 (78.2%), blaTEM (44.9%) and blaSHV (3.8%). The CTX-M variants were also found with the predominance of blaCTX-M-1 (89.7%) and blaCTX-M-15 (89.7%) followed by blaCTX-M-9 (11.5%).
Despite the resistance to most of the tested antibiotics, ExPEC isolates showed fortunately good susceptibility to fosfomycin and carbapenems. blaCTX-M1, blaCTX-M15 and blaOXA-1 seem to be E.coli major ESBL genes circulating in Senegal. The high level of resistance to antimicrobials observed stresses the need of establishing an epidemiological surveillance of antimicrobial resistance in both community and hospital settings.
Keywords: Escherichia coli; Extended spectrum beta-lactamase; Hospital; Dakar-senegal
Abbreviations
AMR: Antimicrobial Resistance; BLI: Beta-Lactamase Inhibitors; CA: Community-Acquired Bacteria; E. coli: Escherichia coli; ESBL: Extended-Spectrum Beta-Lactamases; ExPEC: Extra-Intestinal Pathogenic Escherichia coli; HA: Hospital-Acquired Bacteria; HALD: Aristide le Dantec University Teaching Hospital; MDR: Multidrug Resistant; No-UPEC: No-Uropathogenic Escherichia coli; UPEC: Uropathogenic Escherichia coli; UTI: Urinary Tract Infections
Introduction
Escherichia coli, a common bacteria found in various parts of the human body, is also the predominant bacterial species responsible for Community-Acquired (CA) and Hospital-Acquired (HA) infections at all ages in human [1]. Human pathogenic E. coli strains are classified into two large groups, strains responsible for intestinal infections and those causing extra-intestinal diseases (ExPEC) [2,3].
ExPECs are among the most common Gram-negative bacterial pathogens affecting Human with diverse infections, including Urinary Tract Infections (UTI), bacteremia, meningitis, nosocomial respiratory infections, peritonitis, prostatitis, skin and soft tissue infections [4-6].
In addition, Multidrug Resistant (MDR) ExPECs are now common both in community-acquired and hospital-acquired infections, including resistance to ß-lactams, which are the commonly used antibiotics in human and animal health. The ß-lactams resistance is mainly mediated by production of extended-spectrum betalactamases [7-9]. These enzymes hydrolyze penicillins, cephalosporins (first, second, third, and fourth generation), and monobactams, but are generally inactive against cephamycins and carbapenems. ESBLs are generally inhibited by Beta-Lactamase Inhibitors (BLI) [10,11]. A worrying fact is that mobile genetic elements that harbor ESBL genes also carry others genes conferring resistance to quinolones, aminoglycosides and even carbapenems [12-15].
ESBL-producing ExPECs infections are responsible of extended hospital stays, accompanying high cost and mortality and morbidity [2]. Hence, the importance to establish innovative diagnostic toolkits and performant surveillance system for early detection and monitoring of ExPECs cases, especially in developing countries. In this study, we investigated the antibiotic resistance profile and the ESBL genes carried by ESBL-producting ExPEC isolated at the laboratory of bacteriology laboratory, Aristide le Dantec University Teaching Hospital (HALD) in Dakar, Senegal. Additionally, we compared CA to HA, and uropathogenic E. coli (UPEC) to No- Uropathogenic ExPEC Isolates (No-UPEC).
Material and Methods
Bacterial Isolates
This is a retrospective study and all ExPEC isolates analyzed in this study were collected between January 1st, 2018 and December 31th, 2020 at the Hospital Laboratory of HALD during routine activities and stored at -80°C. Seventy-eight no-duplicate strains were randomly selected from the Laboratory. Strains were isolated from urine (UPEC, n = 51), pus, sputum, bronchial fluid and vaginal samples (no-uropathogenic ExPEC, n = 27). Of the 78 strains, 49 and29 were CA and HA respectively. Culture and Isolation were done based on gold standard microbiological tests and identification by using Api 20E for Enterobacteriaceae (bioMérieux France).
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was performed using the Kirby- Bauer disc diffusion method and results were interpreted according to the committee of the French society of microbiology (CA-SFM, 2020) recommendations. Briefly, bacterial suspensions were prepared at 0.5 McFarland and inoculated onto Mueller-Hinton agar for an overnight incubation at 37°C. These following antibiotic disks were tested: ampicillin (AMP, 10μg), ticarcillin (TIC, 75μg), amoxicillin-clavulanic acid (AMC, 20/10μg), cefalotin (CEF, 30 μg), cefoxitin (FOX, 30 μg), cefotaxime (CTA, 30μg), ceftazidime (CAZ, 30μg), cefepime (CEP, 30μg), aztreonam (AZT, 30μg), imipenem (IMP, 10μg), ertapenem (ERT, 10μg), Nalidixic acid (NAL, 30μg ), ciprofloxacin (CIP, 5 μg), gentamicin (GEN, 10 μg), amikacin (AMI, 30 μg), fosfomycin (FOS, 50 μg), tetracycline (TET, 30 μg) and sulfamethoxazole-trimethoprim (TMS, 1.25μg / 23.75μg). The E. coli ATCC 25922 was used for quality control. ESBL production was appreciated by double-disk synergy test with disks of amoxicillin-clavulanic acid surrounded at a radius of 30 mm by cefepime, ceftriaxone, ceftazidime and aztreonam.
DNA Extraction
Bacterial DNA extraction was performed mechanically by thermal choc. Briefly, a well-separated bacterial colony was dispersed in a tube contained 1ml of sterile distilled water, vortexed, boiled for 15 minutes at 100°C and centrifuged at 13,200 rpm for 10 min. The supernatant was carefully recovered, aliquoted and stored at -20°C until used. To confirm results, extraction was done by Qiagen kit (DNeasy Blood & Tissue Kit (50) Cat. No. / ID: 69504).
ESBL Genes Amplification
A simplex end-point PCR was performed (on Thermocycler 2720, Applied Biosystems, Lincoln Centre Drive, Foster City, California 94404, USA) to detect ESBL genes. Specific primer pairs (Table 1) were used to amplify ESBL genes (blaCTX-M, blaCTX-M-1, blaCTX-M-9, blaCTX-M-15, blaCTX-M-25, blaOXA-1, blaTEM, blaSHV). Each reaction included positive and negative controls. PCRs were carried out in 20 μl reaction volume (2.5 μl DNA + 17.5 μl Master MixFIREPol®). The amplification program consisted of an initial denaturation at 95°C for 3min., 35 PCR cycles (denaturation: 94°C, 30sec., 72°C, 60sec.) and a final elongation at 72°C for 7min. Ten micro liters of each amplicon were separated on 2% agarose gel in 1X TAE buffer for 35 min at 135 volts and the amplified fragment detected using a GelDoc imager (BioRad).
Target genes
Sequences genes
Sizes (bp)
AnnealingTemp (°C)
References
blaCTX-M
F: 5’ - ATGTGCAGYACCAGTAARGTKATGGC - 3’
R: 5’ - TGGGTRAARTARGTSACCAGAAYSAGCGG - 3’592
55
[16]
blaCTX-M-1
F: 5’ - GGTTAAAAAATCACTGCGTC - 3’
R: 5’ - TTACAAACCGTYGGTGACGA - 3’873
50
[16]
blaCTX-M-9
F: 5’ - GTGACAAAGAGAGTGCAACGG - 3’
R: 5’ - ATGATTCTCGCCGCTGAAGCC - 3’856
55
[16]
blaCTX-M-15
F: 5’ - CACACGTGGAATTTAGGGACT - 3’
R: 5’ - GCCGTCTAAGGCGATAAACA - 3’995
50
[16]
blaCTX-M-25
F: 5’ - GCACGATGACATTCGGG - 3’
R: 5’ - AACCCACGATGTGGGTAGC - 3’327
52
[16]
blaOXA-1
F: 5’ - ATGAAAAACACAATACATATC - 3’
R: 5’ - AATTTAGTGTGTTTAGAATGG - 3’830
56
[17]
blaTEM
F: 5’ - TTGGGTGCACGAGTGGGTTA - 3’
R: 5’ - TAATTGTTGCCGGGAAGCTA - 3’506
55
[16]
blaSHV
F: 5’ - TCGGGCCGCGTAGGCATGAT - 3’
R: 5’ - AGCAGGGCGACAATCCCGCG - 3’628
52
[16]
Table 1: Oligonucleotide primers sequence used for PCR to detect ESBL genes.
Statistical Analysis
Statistical analysis and multiple correspondence analysis and data analysis methods were performed with R software. The statistic test used is the Chi-square at 5% risk threshold. P-values are obtained from the proportion comparison test and the level of significance for all statistical tests was set at p < 0.05.
Results
Antibiotic Susceptibility Testing
All the 78 ExPEC isolates were MDR (resistance to at least one drug from at least three classes of antibiotics), and were resistant to ampicillin, ticarcillin, amoxicillin/clavulanic acid combination, cefalotin, cefotaxime, ceftazidime, cefepime and aztreonam (Table 2). Besides, resistance to ciprofloxacin (93.6%, n = 73), tetracycline (91%, n = 71) and sulfamethoxazole-trimethoprim (91%, n = 71) was high, while less frequent for aminoglycosides (gentamicin, 60.3%, n = 47; amikacin, 42.3%, n = 33). In contrast, only 3.8% (n = 3) and 1.3% (n = 1) of the isolates were resistant to ertapenem and fosfomycin respectively, while all were sensitive to imipenem (Table 2). Comparison of resistance profiles between CA and HA, and between UPEC and no-UPEC strain did not show any significant difference, except for ciprofloxacin (Table 2).
Antibiotics
Total strains
Pathogenicity
Origin
Class
Drug
N (%)
UPEC
N (%)No-UPEC
N (%)p
CA
N (%)HA
N (%)p
Beta-lactams
AMP
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
TIC
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
AMC
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
CEF
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
FOX
5 (6.4)
3 (5.9)
2 (7.4)
0.06
4 (8.2)
1 (3.5)
0.06
CTA
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
CAZ
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
CEP
78 (100)
51 (100)
27 (100)
1
49 (100)
29 (100)
1
AZT
70 (89.7)
48 (91.4)
22 (81.5)
0.08
46 (93.9)
24 (82.8)
0.12
IMP
0
0
0
-
0
0
-
ERT
3 (3.8)
3 (5.9)
0
0.01*
2 (4.1)
1 (3.5)
0.04*
Quinolones and Fluoroquinolones
NAL
76 (97.4)
51 (100)
25 (92.6)
0.97
49 (100)
27 (93.1)
0.97
CIP
73 (93.6)
50 (98)
23 (85.2)
0.03*
48 (98)
25 (86.2)
0.04*
Aminoglycosides
GEN
47 (60.3)
33 (64.7)
14 (51.9)
0.27
29 (59.2)
16 (62.1)
0.72
AMI
33 (42.3)
24 (47.1)
9 (33.3)
0.24
19 (38.8)
14 (48.3)
0.41
Phosphonicacid
FOS
1 (1.3)
1 (2)
0
0.01*
0
1 (3.6)
0.01*
Cyclines
TET
71 (91)
45 (88.2)
26 (96.3)
0.24
44 (89.8)
27 (93.1)
0.62
Antifolates
TMS
71 (91)
45 (88.2)
26 (96.3)
0.24
45 (91.8)
26 (89.7)
0.74
UPEC, UropathogenicE. coli; CA, Community-acquired; HA, Hospital-acquired; AMP, ampicillin; TIC, ticarcillin; AMC, Amoxicillin-clavulanic acid; CEF, cefalotin; FOX, cefoxitin; CTA, cefotaxim; CAZ, ceftazidime; CEP, cefepime; AZT, aztreonam; IMP, imipenem; ERT, Ertapenem;NAL, nalidixic acid; CIP, ciprofloxacin; GEN, gentamicin; AMI, amikacin; FOS, fosfomycin; TET, tetracycline; TMS, sulphamethoxazole-trimethoprim; *, significant p-value (p ? 0.05).
Table 2: Antibiotics resistance rate of total strains, CA and HA strains, UPEC and no-UPEC strains
Presence of ESBL Genes
All 78 strains carried at least one ESBL gene. blaCTX-M group was the most prevalent (77/78; 98.7%), followed by blaOXA-1(61/78; 78.2%), blaTEM(35/78; 44.9%) and blaSHV(3/78; 3.8%) (Table 3) and (Figure 1). 51/51 (100%) of UPEC strains and 29/29 (100%) of hospital -acquired strains carried the blaCTX-M gene and none of ‘’ no-uropathogenic ExPEC ‘’ strains carried a blaSHV gene (Table 3) and (Figure 2). (9/78; 11.5%) carried only blaCTX-M or blaOXA-1 and (69/78; 88.5%) carried several types of ESBL gene. Indeed, (2/78;2.6%) carried blaCTXM +blaOXA-1+blaTEM+blaSHV; (23/78; 29.4%) carried blaCTX-M+blaOXA- 1+blaTEM; (1/78; 1.3%) carried blaCTXM+blaOXA-1+blaSHV; (32/78; 41%) carried blaCTX-M+blaOXA-1 and (11/78; 14.1%) of strains carried “blaCTXM +blaTEM” (Table 3). None of strains carried blaTEM or blaSHV gene alone (Table 4).
ESBL
Total strains
Pathogenicity
Origin
Family
Genes
N (%)
UPEC
N (%)No-UPEC
N (%)p
CA
N (%)HA
N (%)p
Cefotaximase-Munich
blaCTX-M
77 (98.7)
51 (100)
26 (96.3)
0.98
48 (98)
29 (100)
0.99
blaCTX-M-1
70 (89.7)
48 (94.1)
22 (81.5)
0.08
44 (89.8)
26 (89.7)
0.98
blaCTX-M-9
9 (11.5)
5 (9.8)
4 (14.8)
0.25
5 (10.2)
4 (13.8)
0.45
blaCTX-M-15
70 (89.7)
48 (94.1)
22 (81.5)
0.08
44 (89.8)
26 (89.7)
0.98
blaCTX-M-25
0
0
0
-
0
0
-
Oxacillinase
blaOXA-1
61 (78.2)
42 (82.4)
19 (70.4)
0.22
38 (77.6)
23 (79.3)
0.86
Temoneira
blaTEM
35 (44.9)
23 (45.1)
12 (44.4)
0.95
19 (38.8)
16 (55.2)
0.16
Sulfhydryl variable
blaSHV
3 (3.8)
3 (5.9)
0
0.01*
2 (4.1)
1 (3.5)
0.04*
UPEC,Uropathogenic E. coli; CA,community-acquired; HA,hospital-acquired; %, percentage; N,number of isolates, *, significant p-value (? 0.05).
Table 3: Prevalence of ESBL genes in total strains, CA and HA strains, UPEC and non-UPEC strains.
Combination of ESBL genes
Total strains
Pathogenicity
Origin
N (%)
UPEC
N (%)No-UPEC
N (%)p
CA
N (%)HA
N (%)p
blaCTX-M-1 + blaCTX-M-15 + blaCTX-M-9 + blaOXA-1 + blaTEM
2 (2.6)
2 (3.9)
0
0.01*
1 (2)
1 (3.4)
0.1
blaCTX-M-1 + blaCTX-M-15 + blaOXA-1 + blaTEM+ blaSHV
2 (2.6)
2 (3.9)
0
0.01*
1 (2)
1 (3.4)
0.1
blaCTX-M-1 + blaCTX-M-15 + blaOXA-1 + blaTEM
21 (26.9)
14 (27.4)
7 (33.3)
0.68
10 (20.4)
11 (37.9)
0.12
blaCTX-M-1 + blaCTX-M-15 + blaOXA-1 + blaSHV
1 (1.3)
1 (2)
0
0.01*
1 (2)
0
0.01*
blaCTX-M-1 + blaCTX-M-15 + blaOXA-1
32 (41)
21 (41.2)
11 (40.7)
0.98
23 (46.9)
9 (31)
0.15
blaCTX-M-1 + blaOXA-1 + blaTEM
1 (1.3)
1 (2)
0
0.01*
1 (2)
0
0.01*
blaCTX-M-1 + blaCTX-M-15 + blaTEM
7 (9)
4 (7.8)
3 (11.1)
0.21
5 (10.2)
2 (6.9)
0.23
blaCTX-M-1 + blaCTX-M-15
3 (3.8)
2 (3.9)
1 (3.7)
0.97
2 (4.1)
1 (3.4)
0.04
blaCTX-M-15 + blaCTX-M-9
2 (2.6)
2 (3.9)
0
0.01*
1 (2)
1 (3.4)
0.1
blaCTX-M-1 + blaOXA-1
1 (1.3)
1 (2)
0
0.01*
0
1 (3.4)
0.01*
blaCTX-M-9 + blaTEM
2 (2.6)
0
2 (7.4)
0.01*
1 (2)
1 (3.4)
0.1
UPEC, Uropathogenic E. coli; CA, community-acquired; HA, hospital-acquired %, percentage; N, number of isolates; *, significant p-value (? 0.05).
Table 4: Prevalence of ESBL genes combinations in total strains, CA and HA strains, UPEC and non-UPEC strains.
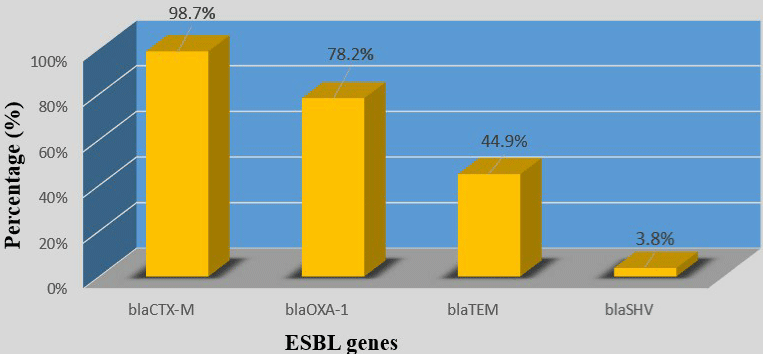
Figure 1: Prevalence of ESBL genes in total strains.
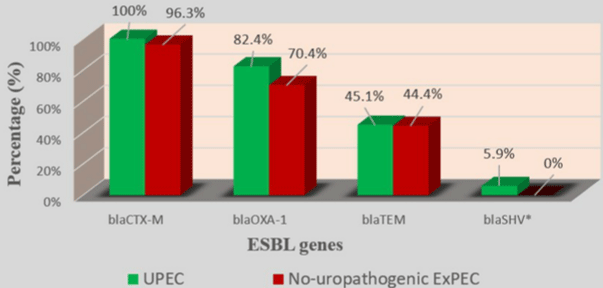
Figure 2: Prevalence of ESBL genes in UPEC and no-uropathogenic ExPEC.
*Significant p (p = 0.01)
In 4 blaCTX-M group, blaCTX-M-1 (70/78; 89.7%) with blaCTX-M-15 (70/78; 89.7%) was the most prevalent followed by blaCTX-M-9 (9/78; 11.5%). blaCTX-M-25 was not detected in any of the 77 strains (Table 4). Among strains which carried the blaCTX-M type, 89.6% carried 2 variants of blaCTX-M while 7.8% carried only one variant and 2.6% carried 3 blaCTX-M variants (Table 4). No significant difference was found by comparing the prevalence of ESBL genes in hospital-acquired and communityacquired strains on the one hand and UPEC and no-uropathogenic ExPEC strains on the other hand (Figures 2-5).
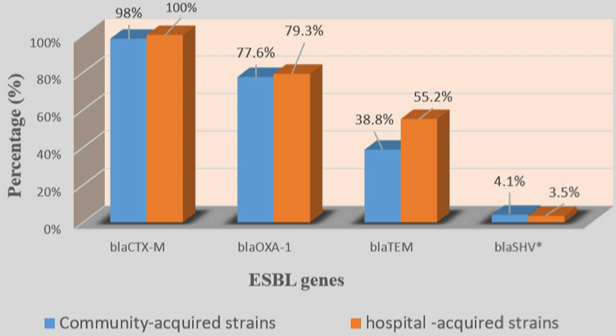
Figure 3: Prevalence of ESBL genes in community-acquired and hospital-acquired strains.
*Significant p (p = 0.04)
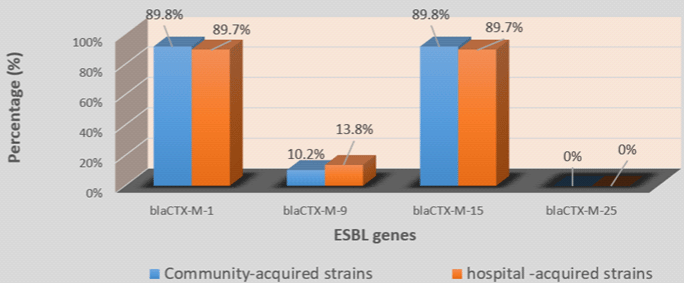
Figure 4: Prevalence of blaCTX-M variants in community and hospital-acquired strains.
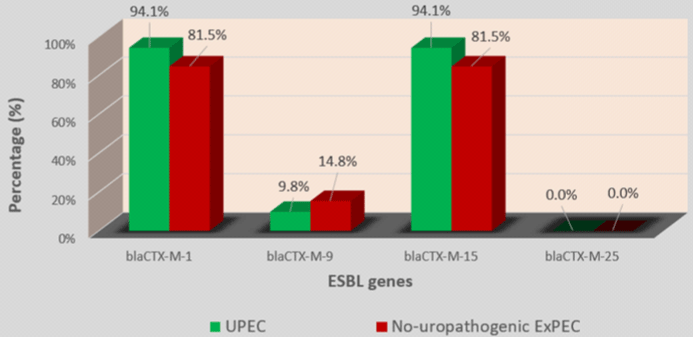
Figure 5: Prevalence of blaCTX-M variants genes in UPEC and no-uropathogenic ExPEC.
Discussion
Potentially pathogenic Escherichia coli which produce Extended Spectrum Betalactamases (ESBL) are frequently isolated from urinary tract infections [6-18] can be resistant to many molecules of this class. The most worrying thing is that these strains, which spread rapidly both in hospitals and in the community, are often resistant to many other antibiotics such as those of the aminoglycosides and quinolones classes, thus making the treatment failure of these infections.
The rate of resistance to multiple antibiotics among ESBLproducing isolates is usually common due to carrying multi- resistant genes and plasmids [12-14].
Nowadays, ESBL type CTX-M are the most widespread in the world, unlike ESBL TEM and SHV which are becoming less prevalent [19,20]. Our study also confirmed this trend with 98.7% of strains positive for blaCTX-M, followed by blaOXA-1 (78.2%), blaTEM (44.9%) and blaSHV (3.8%). Several studies carried out in Togo [21], in Saudi Arabia [22], and in Mozambique [23] reported high prevalence rates of blaCTX-M in ESBL ExPEC strains 100%; 93,94% and 77%, respectively. Moreover, other authors [24,25] had already pointed out that currently, blaOXA-1 was the second most prevalent ESBL gene type in the world behind blaCTX-M. In disagreement to these studies, [23] rather mentioned 52% of prevalence for blaSHV and 1% for blaTEM in 2021 in Mozambique. [26] mentioned 3.12% of prevalence rate for blaSHV in 2019 in Senegal. ESBLs SHV-gene type therefore seems to be rare in E. coli strains circulating in Senegal.
Interestingly, 55.1% of the strains harbored 2 ESBL gene types while 30.8% of the strains carried 3 and 2.6% carried all the 4 ESBL genotypes. While 33.33% and 12.12% of strains carrying 2 and 3 ESBL gene types, respectively were reported from Riyadh, Saudi Arabia [22]. The very high proportion of strains (88.5%) combining several ESBL gene types seems to be one of the major causes of the 100% resistance to ampicillin, ticarcillin, (clavulanic acid + amoxicillin), cefalotin, cefotaxime, ceftazidime, cefepime and aztreonam.None of the strains carried only blaTEM or blaSHV gene.
Globally, blaCTX-M-15 had long been cited as the most prevalent variant of blaCTX-M in E. coli [27-29]. The high prevalence rate of blaCTX-M-15 (89.7%) observed among blaCTX-M positive strains in our study corroborates these earlier studies. An interesting fact in our study was that blaCTX-M-1 was as prevalent as blaCTX-M-15 with respectively 89.7% and 85.7% rates of the strains concomitantly carried blaCTX-M-15 and blaCTX-M-1. These data suggest that blaCTX-M-15 is not the only major variant of blaCTX-M circulating in Senegal. The low prevalence rates of blaCTX-M-9 (11.5%) and blaCTX-M-25 (0%) follow trends observed in other parts of the world [27-30].
No significant difference was noted when comparing the prevalence of ESBL genes from community-acquired and hospitalacquired strains. This seems to imply either a port of ESBL ExPEC in community or that the community strains are the same ones encountered in a hospital environment. We did not notice any significant difference in the prevalence of ESBL genes between UPEC strains and non-uropathogenic ExPEC strains. It seems that in Senegal, non-uropathogenic ExPEC are as resistant as UPEC strains. Future studies could confirm this.
The high prevalence of blaCTX-M genes suggests the involvement of mobile genetic elements (plasmids, integrons and transposons) [20,31] in the spread of antibiotic resistance in Dakar, as reported in many studies leading increasing resistance to fluoroquinolones, aminoglycosides and even carbapenems antibiotics [12-14]. This suggests the importance to study and monitor the mobile genetic elements from strains isolated in healthy carriers, environment, and hospital settings in order to initiate others actions that can help fighting against antibiotic resistance.
Conclusion
All the 78 ExPEC strains tested in this study were MDR patterns, and resistant to almost all antibiotics families, except fosfomycin and carbapenems. Based on our results, we recommend avoiding monotherapy and prohibiting fluoroquinolones, C3G and C4G as empiric treatment of UTIs in Senegal. blaCTX-M (blaCTX-M1, blaCTX-M15) and blaOXA-1 seem to be the major ESBL genes circulating in Senegal. No significant difference was noted when comparing the prevalence of ESBL genes between hospital-acquired and community-acquired strains; As well as by comparing UPEC and ExPEC strains isolated from other types of samples. The high resistance to antimicrobials and extended virulence factors profiles observed, underscore the relevance to implement an epidemiological Antimicrobial Resistance (AMR) surveillance system to improve the management of treatment protocols in patients infected with MDR bacteria.
Ethical Research Approval
This study has received the Ethical Research approval of the Research Ethics Committee (CER) of Cheikh Anta Diop University (UCAD) under the reference CER/UCAD/AD/MSN/051/2020.
Authors Contributions
CM was the main project administrator, designed this research project, supervised entire research and revised manuscript. SBB designed this research project, supervised entire research and revised manuscript.FK, DSH, DA, TA, NSML and NCS Contributed to collect samples. CA and NI helped to perform laboratory works. DY, FC and SA supervised research and revised manuscript. LG, BDA, DNH, TKC, GDA, MS and BCSB served as resource scientists during this project. DKM designed this research project, drafted the manuscript, perfomed laboratory works and analyzed and interpreted results.
Conflict of Interest
The authors have not declared any conflict of interests.
Acknowledgements
The authors thank Abdoul Aziz Wane, Amadou Mactar Gueye, Ousmane Sow Brice Leon Mosso and El Hadji Aly Niang for their technical assistance.
References
- Savage DC. Microbial biota of the human intestine: a tribute to some pioneering scientists. Current issues in intestinal microbiology. 2001; 2: 1-15.
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Reviews Microbiology. 2004; 2: 123-140.
- Braz VS, Melchior K, Moreira CG. Escherichia coli as a Multifaceted Pathogenic and Versatile Bacterium. Frontiers in Cellular and Infection Microbiology. 2020; 10.
- Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes and infection. 2003; 5: 449-456.
- Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli : “The other bad E coli.” J Lab Clin Med. 2002; 139: 155–162.
- Pitout JDD. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Frontiers in Microbiology. 2012; 3.
- Camara M, Mane MT, Ba-Diallo A, Dieng A, Diop-Ndiaye H, Karam F, et al. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae clinical isolates in a Senegalese teaching hospital: A cross sectional study. Afr J Microbiol Res. 2017; 11: 1600–1605.
- Toudji AG, Djeri B, Karou SD, Tigossou S, Ameyapoh Y, Souza C de. Prévalence des souches d’entérobactéries productrices de bêta-lactamases à spectre élargi isolées au Togo et de leur sensibilité aux antibiotiques. Int J Biol Chem Sci. 2017; 11: 1165–1177.
- Ouedraogo A, Sanou M, Kissou A, Sanou S, Solaré H, Kaboré F, et al. High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infectious Diseases. 2016; 16.
- Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended Spectrum Betalactamases: Definition, Classification and Epidemiology. Current issues in molecular biology. 2015; 17: 11-21.
- Bradford PA. Extended-Spectrum ß-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clinical Microbiology Reviews. 2001; 14: 933-951.
- Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. The Journal of antimicrobial chemotherapy. 2012; 67:346-356.
- Doumith M, Day M, Ciesielczuk H, Hope R, Underwood A, Reynolds R, et al. Rapid Identification of Major Escherichia coli Sequence Types Causing Urinary Tract and Bloodstream Infections. Journal of Clinical Microbiology. 2014; 53: 160-166.
- Dale AP, Woodford N. Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. The Journal of infection. 2015; 71: 615-626.
- Poirel L, Dortet L, Bernabeu S, Nordmann P. Genetic Features of blaNDM-1- Positive Enterobacteriaceae. Antimicrobial Agents and Chemotherapy. 2011; 55: 5403-5407.
- Gundran RS, Cardenio PA, Villanueva MA, Sison FB, Benigno CC, Kreausukon K, et al. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended- spectrum ß- lactamase- producing E. coli isolates from broiler farms in the Philippines. BMC Veterinary Research. 2019; 15.
- Weill F, Guesnier F, Guibert V, Timinouni M, Demartin M, Polomack L, et al. Multidrug Resistance in Salmonella enterica Serotype Typhimurium from Humans in France (1993 to 2003). Journal of Clinical Microbiology. 2006; 44: 700-708.
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005; 18: 657–686.
- Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M ß-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017; 72: 2145–2155.
- Cantón R, González-Alba JM, Galán JC. CTX-M Enzymes: Origin and Diffusion. Frontiers in Microbiology. 2012; 3.
- Dossim S, Salou M, Ihou-Wateba M, Bidjada B, Godonou AM, Aoussi E, et al. Molecular Characterization Of Extended Spectrum Beta lactamases Genes (Blactxm And Blashv) In Enterobacteria Isolates In Medical Specimens In Lomé (Togo).
- Alqasim A, Jaffal AA, Alyousef AA. Prevalence of Multidrug Resistance and Extended-Spectrum ß-Lactamase Carriage of Clinical Uropathogenic Escherichia coli Isolates in Riyadh, Saudi Arabia. International Journal of Microbiology. 2018; 2018: 1-9.
- Estaleva CEL, Zimba TF, Sekyere JO, Govinden U, Chenia HY, Simonsen GS, et al. High prevalence of multidrug resistant ESBL- and plasmid mediated AmpC-producing clinical isolates of Escherichia coli at Maputo Central Hospital, Mozambique. BMC Infect Dis. 2021; 21: 16.
- Faezi Ghasemi M, Dibadji SN. Prevalence of blaoxa-1 and blashv Genes in E. coli Isolates from Hospitalized Patients in Rasht. Med Lab J. 2016; 10: 65–70.
- Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of blaCTX - M, blaTEM, blaSHV and blaOXA genes in Extended-spectrum-ß- lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrobial Resistance and Infection Control. 2019; 8.
- Diagne R. Recherche de gènes BLSE de type TEM, SHV, et OXA-1 sur des souches de E. coli isolées au laboratoire de Bactériologie de Fann, Sénégal. Rev Afr Malgache Rech Sci Santé. 2019; 1.
- Lahlaoui H, Khalifa ABH, Moussa MB. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum ß-lactamase (ESBL). Medecine et maladies infectieuses. 2014; 44: 400-404.
- Moghaddam MN, Beidokhti MH, Jamehdar SA, Ghahraman M. Genetic properties of blaCTX-M and blaPER ß-lactamase genes in clinical isolates of Enterobacteriaceae by polymerase chain reaction. Iranian Journal of Basic Medical Sciences. 2014; 17: 378-383.
- Zong Z, Partridge SR, Thomas L, Iredell JR. Dominance of blaCTX-M within an Australian Extended-Spectrum ß-Lactamase Gene Pool. Antimicrobial Agents and Chemotherapy. 2008; 52: 4198-4202.
- Pavez M, Troncoso C, Osses I, Salazar R, Illesca V, Reydet P, et al. High prevalence of CTX-M-1 group in ESBL-producing enterobacteriaceae infection in intensive care units in southern Chile. The Brazilian journal of infectious diseases : an official publication of the Brazilian Society of Infectious Diseases. 2019; 23: 102-110.
- Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. International journal of medical microbiology. IJMM. 2011; 301: 654-658.