
Research Article
Austin J Microbiol. 2024; 9(2): 1050.
Analysis of Pancreatic Cancer Intestinal Tissue Microbiota Structure and Pathogens Based on Microbiome Data
Hao Dong¹; Yuqi Wang¹; Linlin Lu²*; Xuezhen Ma³*
1College of Medicine, Qingdao University, Qingdao 266071, China
2Qingdao Cancer Prevention and Treatment Research Institute, Qingdao Central Hospital, University of Health and Rehabilitation Sciences (Qingdao Central Medical Group), Qingdao 266042, China
3Cancer Center, Qingdao Central Hospital, University of Health and Rehabilitation Sciences (Qingdao Central Medical Group), Qingdao 266042, China
*Corresponding author: Xuezhen Ma Cancer Center, Qingdao Central Hospital, University of Health and Rehabilitation Sciences (Qingdao Central Medical Group), Qingdao 266042, China; Linlin Lu, Qingdao Cancer Prevention and Treatment Research Institute, Qingdao Central Hospital, University of Health and Rehabilitation Sciences (Qingdao Central Medical Group), Qingdao 266042, China, Email: 18660229289@163.com; 18266208011@163.com
Received: March 19, 2024 Accepted: April 24, 2024 Published: May 01, 2024
Abstract
Objective: This study aims to investigate the impact of pancreatic cancer on the gut microbiota structure in adults by analyzing the diversity and composition changes of microbes, and to explore the relationship between pancreatic cancer and intestinal microbiota.
Results: The study demonstrated significant differences in the microbiota structure between pancreatic cancer patients and healthy individuals. Changes in microbial diversity and composition were confirmed through a-diversity and β-diversity analyses. A random forest model presented the gut-related microbial community patterns. The Microbial Infection Potential (MIP) indicated variations in pathogen numbers under different conditions.
Methods: Fecal samples from healthy individuals and pancreatic cancer patients were collected for microbial community structure comparison using 16S rRNA gene sequencing. The changes in microbial composition and pathogen characteristics were revealed through a-diversity and β-diversity analyses, as well as metagenomic analysis.
Conclusion: This study not only elucidated the impact of pancreatic cancer on intestinal microbiota but also suggested its potential role in the development of pancreatic cancer. Understanding these interactions may help discover new biomarkers, therapeutic targets, and personalized treatment strategies. Future research will explore the clinical application potential of these microbiomes in the treatment of pancreatic cancer and further reveal underlying mechanisms.
Keywords: Molecular Biology; Microbiome; Pancreatic Neoplasm
Introduction
Pancreatic cancer, a devastating malignancy with a poor prognosis, has been increasingly linked to alterations in the gut microbiota. The intricate relationship between pancreatic cancer and the intestinal microbiome is gaining attention for its potential role in cancer development, progression, and treatment response. This paper aims to delve into the complexities of this relationship by analyzing the microbiota structure and identifying key pathogens in pancreatic cancer patients.
The gut microbiota, a complex ecosystem of microorganisms, plays a crucial role in human health, influencing metabolism, immunity, and even the efficacy of cancer therapies [1,2]. In pancreatic cancer, the microbiota undergoes significant changes, characterized by reduced microbial diversity and an increase in pathogenic bacteria [3]. These dysbiotic alterations can trigger persistent inflammation, modulate the tumor microenvironment, and potentially contribute to carcinogenesis [4,5].
Recent studies have highlighted the distinct gut microbiome profiles in pancreatic cancer patients, with a decrease in butyrate-producing bacteria and an increase in harmful genera like Fusobacterium, Enterobacter, and Enterococcus [6]. This shift in microbial composition may influence the host's immune response and increase the risk of cancer development [7]. Furthermore, the microbiota's role extends to affecting the outcome and survival of patients, particularly those undergoing immunotherapy [8].
The interaction between pancreatic cancer and the gut microbiota is bidirectional. Pancreatic factors, such as antimicrobial excretion, can impact the composition and functional properties of the gut microbiota [9]. Moreover, an altered oral microbiota may colonize the pancreas, causing local inflammation and contributing to cancer development [10].
Understanding the correlation between gut microbiota and pancreatic cancer could aid in diagnosis, treatment, and the development of new therapeutic strategies, including the use of probiotics, antibiotics, and fecal microbiota transplantation [11]. This study aims to further elucidate these interactions, offering insights into the potential role of intestinal microbiota in the development and treatment of pancreatic cancer.
Results
Sample Description
This paper studied 180 human intestinal microbiome samples, including 74 samples from healthy adults, 88 samples from patients with pancreatic cancer, and 18 samples from adults in the early stages of pancreatic cancer. All samples were sourced from the NCBI public open-access database.
Microbial Diversity in Healthy Samples, Pancreatic Cancer Samples, and Early-Stage Pancreatic Cancer Samples
After a series of processing steps, this study obtained 180 samples from three different states: healthy young individuals, pancreatic cancer patients, and early-stage pancreatic cancer patients, amounting to a total of 960,210 high-quality sequences. Among these, 74 samples were from healthy young individuals, 88 from pancreatic cancer patients, and 18 from non-tumor-invaded individuals. Species resolution of sequencing data was performed using the Greengenes 13-8 reference database, resulting in 10,669 OTUs. Venn diagram (Figure 1a) showed that the healthy group and pancreatic cancer group shared 4,259 OTUs (39.92%), the healthy group and early-stage pancreatic cancer shared 110 OTUs (1.03%), and the pancreatic cancer group and early-stage pancreatic cancer shared 206 OTUs (1.93%), with a total of 1,171 OTUs (10.98%) shared among all three states. The pancreatic cancer group had 2,311 unique OTUs (21.66%), the healthy group had 2,000 unique OTUs (18.75%), and the early-stage pancreatic cancer group had 612 unique OTUs (5.73%). Twelve phyla were identified across the healthy, pancreatic cancer, and early-stage groups, with Proteobacteria being the dominant phylum in all three groups (45.10%, 37.10%, and 47.96% respectively), followed by Firmicutes (25.16%, 28.80%, and 28.91%). A total of 100 genera were detected in the healthy and pancreatic cancer groups, and 93 genera in the early-stage pancreatic cancer group. Prevotella was the dominant genus in both the healthy and pancreatic cancer groups (20.15% and 18.70% respectively). Pseudomonas was the dominant genus in the early-stage pancreatic cancer group (40.59%), and the second most prevalent genus in the pancreatic cancer group (13.26%). Streptococcus was the second most prevalent genus in both the healthy and early-stage pancreatic cancer groups (15.46% and 13.93%).

Figure 1: Microbial diversity in healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples. A) Venn diagram showing species common and unique among the three groups. B) Comparison of a-diversity (Simpson index and Shannon index) among the three groups. C) Comparison of β-diversity between the healthy samples and pancreatic cancer samples.
Regarding a-diversity, Chao1, Shannon, and Simpson indices were calculated to assess significant differences between the two groups. At the phylum level, both Shannon and Simpson indices showed significant differences between the groups (Figure 1b), with gender having little impact on the results. The a-diversity indicated that the bacterial diversity of the pancreatic cancer group was higher than that of the healthy group. In terms of β-diversity (Figure 1.c), based on Euclidean distance, there was a significant difference in the microbial composition between the healthy group and the pancreatic cancer group (permutations = 999, Pseudo-F = 2.728, R2 = 0.05901, P = 0.00015).
Pathogen Composition in Healthy Samples, Pancreatic Cancer Samples, and Early-Stage Pancreatic Cancer Samples
Regarding pathogens, 13 phyla were identified in the healthy, pancreatic cancer, and early-stage pancreatic cancer groups. In the healthy and pancreatic cancer groups, Firmicutes was the most abundant phylum, accounting for 53.47% and 55.43% of the total abundance, respectively, while Proteobacteria was the second most abundant, constituting 27.41% and 22.68%. In the early-stage pancreatic cancer group, Proteobacteria was the most abundant phylum, representing 47.22% of the total abundance, and Firmicutes was the second most abundant at 27.64%. These results indicate that at the phylum level, the microbial community compositions of the healthy, pancreatic cancer, and early-stage groups are similar, but there are slight differences in abundance, and a significant change in the abundance of Proteobacteria during the progression of pancreatic cancer. At the genus level, the composition of pathogenic microbial communities differed from the overall microbial community composition in all three groups. Eighty-four genera were identified in both the healthy and pancreatic cancer groups, while seventy-seven genera were identified in the early-stage group. Bacillus was the most abundant genus in both the healthy and pancreatic cancer groups, comprising 42.12% and 41.29% of the total abundance, respectively. Ralstonia was the second most abundant genus in these groups, accounting for 8.93% and 8.21%. In the early-stage group, Pseudomonas was the most abundant genus, constituting 41.10% of the total abundance, while Streptococcus was the second most abundant at 13.20%. These findings suggest that at the genus level, the composition of the microbial communities in the three groups differs from the overall microbial community composition, and there may be an increase in the abundance of Pseudomonas and Streptococcus during the progression of pancreatic cancer, which might be associated with the development of the disease.
For a-diversity, Chao1, Shannon, and Simpson indices were calculated to assess community diversity and richness. At the phylum level, Shannon and Simpson indices indicated significant differences between the groups (Figure 2a), suggesting that the bacterial diversity of the pancreatic cancer group was higher than that of the healthy group. In terms of β-diversity (Figure 2b), based on Euclidean distance, there was a significant difference in bacterial composition between the healthy group and the pancreatic cancer group (permutations = 999, Pseudo-F = 5.184, R2 = 0.10648, P = 0.00092).
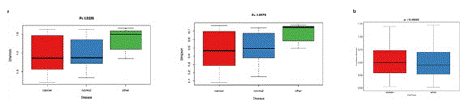
Figure 2: A) Microbial diversity and structure of the pathogen community in healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples. Comparison of a-diversity (Simpson index and Shannon index) among the three groups. B) Comparison of β-diversity in the pathogen community between healthy samples and pancreatic cancer samples.
Risk Level of Microbial Communities
The Microbial Infection Potential (MIP) can be used to assess the overall risk level of microbial communities [12]. This index was used to compare the risk levels of opportunistic pathogens in healthy samples and pancreatic cancer samples. The results showed that there were no significant differences in the risk levels of opportunistic pathogens between the healthy and pancreatic cancer groups. However, the early-stage pancreatic cancer group differed from both (Figure 3a). In terms of oral and ENT (ear, nose, throat) risks, there were no significant differences between the healthy and pancreatic cancer groups, but the risk level in the early-stage pancreatic cancer group was higher than in the healthy and pancreatic cancer groups. This indicates that the progression of pancreatic cancer can greatly affect the risk of opportunistic pathogen infections in the human oral cavity and ENT areas. A similar phenomenon was observed in the skin and circulatory system (Figure 3b).
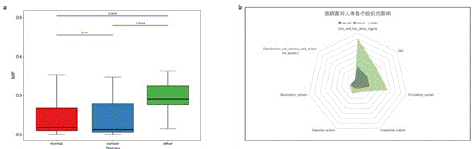
Figure 3: Overall risk level scoring of the microbial community for all samples. A) Box plot of the final MIP scores for healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples. B) Radar chart of MIP scores in various human organ systems for healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples.
Classification of Health Status Using Bacterial Biomarkers
This study established a random forest classifier for specifically identifying healthy samples from pancreatic cancer samples. The top 30 most abundant species were selected to construct the classification model. The classifier's cross-validation error curve was obtained through 10 iterations of tenfold cross-validation. At an average accuracy requirement of >80% (Figure 4a), 2 genera were identified as the best marker set for distinguishing healthy, pancreatic cancer, and early-stage pancreatic cancer samples. Additionally, when the average accuracy rate was set to >80% (Figure 4b), 1 pathogenic genus marker was identified as the optimal marker set for distinguishing between healthy, pancreatic cancer, and early-stage pancreatic cancer samples.
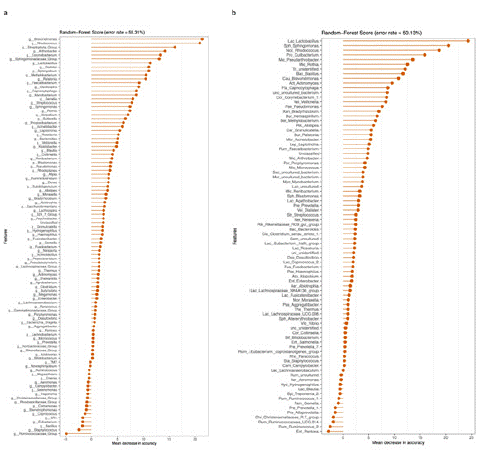
Figure 4: Random forest importance evaluation at the genus level. A) Healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples. B) Differentiation of healthy samples, pancreatic cancer samples, and early-stage pancreatic cancer samples in the pathogen community.
LEfSe Microbiome Difference Analysis in Healthy Samples and Pancreatic Cancer Samples
The LEfSe method was used to analyze microbiome differences in the samples. As shown in Figure 5, the analysis revealed that Serratia was the most contributing differential species in the healthy sample group, while Arthrobacter was the most contributing in the pancreatic cancer sample group (Figure 5a). Notably, Dialister, often associated with clinical infections, was prominently increased in the pancreatic cancer group. Figure 5.a clearly shows that pancreatic cancer leads to a significant decrease in beneficial microbes in the human body, while increasing harmful microbes such as Dialister. Furthermore, Figure 5.b displays the evolutionary phylogenetic relationships of the differential species, where the circles radiating from the inside out represent taxonomic levels from phylum to genus. It's notable that the pancreatic cancer group exhibited differences at the family taxonomic level.
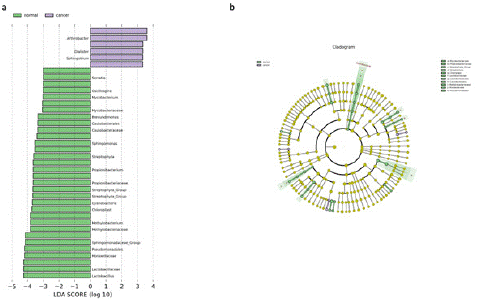
Figure 5: LEfSe microbiome difference analysis for healthy samples and pancreatic cancer samples. A) LDA value distribution chart for different species in healthy samples and pancreatic cancer samples, where color represents the corresponding group and the length of the bar represents the contribution of different species. B) Species evolutionary branching diagram for different species in healthy samples and pancreatic cancer samples, with concentric circles radiating from the inside out representing taxonomic levels from phylum to genus. Each small circle at different taxonomic levels represents a category at that level, with the diameter of the circle proportional to relative abundance.
Methods
Sample Data
Due to the particularity of pancreatic cancer and the emotions of patients and their families, this study used real data already published in public databases for analysis. The original sequences and sampling information came from the publicly available NCBI project: PRJNA832909.
Bioinformatics Analysis
For sequences converted to fastq format, this study employed FastQC and Trim Galore software to filter out sequences with low scores and short lengths, maintaining a Phred score threshold of 20 and a length threshold of 100 nucleotides. Quality-controlled paired sequences were merged using FLASH, retaining sequences with a merging rate of over 80%, and non-biological sequences were removed using VSEARCH. Downstream bioinformatics analysis was conducted using mothur. High-quality sequences were clustered into ASVs with a 99% identity threshold and annotated using the Greengenes 13-8 reference database.
Statistical Analysis
Before calculating diversity indices, all samples underwent sequence rarefaction due to significant differences in sequence counts among different samples. This study calculated a-diversity metrics, including the Shannon, Chao1, and Simpson indices. β-diversity differences were computed using Euclidean distance and tested through Permutational Multivariate Analysis of Variance (PERMANOVA). When studying the impact of pancreatic cancer on intestinal microbiota, the MIP was used to assess whether pancreatic cancer could cause changes in the quantity and proportion of potential pathogens in the intestinal microbiota. The advantage of this method is that it allows for a more specific understanding of the impact of pancreatic cancer on the intestinal microbiota, rather than just assessing differences in microbial diversity and composition. MIP can more accurately assess the pathogenicity of the intestinal microbiota, taking into account the abundance and frequency of various pathogens, and further reporting their targeted human organs or body parts, such as the Oral and Five Sense Organs (OFSO), Skin, Circulatory System (CS), Urogenital System (US), Digestive System (DS), Respiratory System (RS), and other systems (e.g., musculoskeletal, nervous, and endocrine systems).
Conclusion
This study underscores the significant impact of pancreatic cancer on the intestinal microbiota, revealing distinct changes in microbial diversity and composition. Pancreatic cancer leads to reduced microbial diversity in the intestine, with increased species like Veillonella, Klebsiella, and LPS-producing bacteria, altering the gut microbiome composition and potentially influencing tumor aggressiveness and the microenvironment [4,7,13]. These alterations in the gut microbiota are not only indicative of the disease state but may also play a role in the development and progression of pancreatic cancer by stimulating persistent inflammation, regulating the immune system, and modifying the tumor microenvironment [14].
Moreover, our findings suggest that the gut microbiota's composition and metabolic pathways are distinctly altered in pancreatic cancer patients, which could be pivotal in understanding the disease's pathogenesis and progression [15]. The Microbial Infection Potential (MIP) in our study indicated variations in pathogen numbers, highlighting the role of gut microbiota in modulating the inflammatory response and producing carcinogenic metabolic products [16,17].
In conclusion, the intricate relationship between pancreatic cancer and intestinal microbiota opens new avenues for biomarker discovery, therapeutic targets, and personalized treatment strategies. Future research should focus on the clinical application potential of these microbiomes in treating pancreatic cancer and further elucidating the underlying mechanisms.
References
- Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017; 402: 9-15.
- Zhang X, Liu Q, Liao Q, Zhao Y. Pancreatic Cancer, Gut Microbiota, and Therapeutic Efficacy. J Cancer, 2020; 11: 2749-2758.
- Merali N, Chouari T, Terroire J, Jessel MD, Liu DSK, Smith JH, et al. Bile Microbiome Signatures Associated with Pancreatic Ductal Adenocarcinoma Compared to Benign Disease: A UK Pilot Study. Int J Mol Sci. 2023; 24: 16888.
- Sobocki B K, Kazmierczak-Siedlecka K, Folwarski M, Hawrylkowicz V, Makarewicz W, Stachowska E. Pancreatic Cancer and Gut Microbiome-Related Aspects: A Comprehensive Review and Dietary Recommendations. Nutrients. 2021; 13: 4425.
- Ji Zhouxin, He De. Microbiota and pancreatic cancer. International Journal of Oncology. 2020: 46-50.
- Wang X, Sun Z, Makale EC, et al. Gut Microbial Profile in Patients with Pancreatic Cancer. Jundishapur J Microbiol. 2022; 15: e122386.
- Archibugi L, Signoretti M, Capurso G. The Microbiome and Pancreatic Cancer: An Evidence-based Association?. J Clin Gastroenterol. 2018; 52: S82-s85.
- Abdul Rahman R, Lamarca A, Hubner RA, Valle JW, McNamara MG, et al. The Microbiome as a Potential Target for Therapeutic Manipulation in Pancreatic Cancer. Cancers (Basel). 2021; 13: 3779.
- Adolph TE, Mayr L, Grabherr F, Schwarzler J, Tilg H. Pancreas–Microbiota Cross Talk in Health and Disease. Annual Review of Nutrition. 2019; 39: 249-266.
- Sammallahti H, Kokkola A, Rezasoltani S, Ghanbari R, Aghdaei HA, Knuutila S, et al. Microbiota Alterations and Their Association with Oncogenomic Changes in Pancreatic Cancer Patients. Int J Mol Sci. 2021; 22: 2978.
- Li Q, Jin M, Liu Y, Jin L. Gut Microbiota: Its Potential Roles in Pancreatic Cancer. Front Cell Infect Microbiol. 2020; 10: 572492.
- Sun Z, Liu X, Jing G, Chen Y, Jiang S, Zhang M, et al. Comprehensive understanding to the public health risk of environmental microbes via a microbiome-based index. J Genet Genomics. 2022; 49: 685-688.
- Bangolo A I, Trivedi C, Jani I, Pender S, Khalid H, Alqinai B, et al. Impact of gut microbiome in the development and treatment of pancreatic cancer: Newer insights. World J Gastroenterol. 2023; 29: 3984-3998.
- Chen Z, Zhang S, Dong S, Xu H, Zhou W. Association of the Microbiota and Pancreatic Cancer: Opportunities and Limitations. Front Immunol. 2022; 13: 844401.
- Half E, Keren N, Reshef L, Dorfman T, Lachter I, Kluger Y, et al. Fecal microbiome signatures of pancreatic cancer patients. Sci Rep. 2019; 9: 16801.
- Al-Qadami GH, Secombe KR, Subramaniam CB, Wardill HR, Bowen JM. Gut Microbiota-Derived Short-Chain Fatty Acids: Impact on Cancer Treatment Response and Toxicities. Microorganisms. 2022; 10.
- Zhang W, An Y, Qin X, Wu X, Wang X, Hou H, et al. Gut Microbiota-Derived Metabolites in Colorectal Cancer: The Bad and the Challenges. Front Oncol. 2021; 11: 739648.