
Review Article
J Mol Biol & Mol Imaging. 2014;1(2): 8.
Transfer RNA as a Source of Small Functional RNA
Megumi Shigematsu, Shozo Honda and Yohei Kirino*
Computational Medicine Center, Department of Biochemistry and Molecular Biology, Sidney Kimmel Medical College, Thomas Jefferson University, USA
*Corresponding author: Yohei Kirino, Computational Medicine Center, Department of Biochemistry and Molecular Biology, Sidney Kimmel Medical College, Thomas Jefferson University, USA
Received: September 09, 2014; Accepted: October 04, 2014; Published: October 08, 2014
Abstract
Since their discovery in the 1950s, transfer RNAs ( tRNAs ) have been best known as adapter molecules that play a central role in translating genetic information. However, recent biochemical and bioinformatics evidence has led to a previously unexpected conceptual consensus that tRNAs are not always end products; they further serve as a source of small functional RNAs. In many organisms, specific tRNA fragments are produced from mature tRNAs or their precursor transcripts not as random degradation products, but as functional molecules involved in many biological processes beyond translation. In this review, we summarize recent studies of tRNA fragments that have provided new insights into tRNA biology by examining the molecular functions of tRNA fragments and proteins with which they interact.
Keywords: tRNA; tRNA fragment; tRNA half; tRF; Argonaute
Introduction
The groundbreaking development of next-generation sequencing (NGS) technologies has dramatically advanced our understanding of the cellular transcriptome, revealing that non-protein-coding regions of the genome are widely transcribed, and the generated non-coding RNAs (ncRNAs) play important roles in normal biological processes and diseases [1]. Within the diverse group of ncRNAs, the functional significance is particularly evident for small regulatory RNAs which direct the highly-specific regulation of gene expression by recognizing their complementary RNA targets [2-4]. To date, the following three major classes of small regulatory RNAs have been reported: micro RNAs (miRNAs), short-interfering RNAs (siRNAs), and PIWIinteracting RNAs (piRNAs). The definitive features of these RNAs are their short lengths of 20-31 nucleotides (nt) and their interaction with Argonaute family proteins to form effectors ribonucleoprotein complexes. miRNAs, the best-studied class of small regulatory RNAs, repress complementary target mRNA expression, which is estimated to regulate the expression of most protein-coding genes [5, 6]. Thus, small regulatory RNAs constitute one of the most abundant gene expression regulators and exhibit a tremendous impact on all biological processes by shaping the transcriptome. Although such small RNAs have been the focus of much attention over the recent years, NGS studies combined with RNA biochemical studies have revealed the existence of many different other functional small ncRNAs in the cellular transcriptom'e, including small RNA fragments from transfer RNAs (tRNAs), which we will highlight in this review.
Unexpected Expansion of the tRNA World
tRNAs are universally expressed in all three domains of life, and play a central role in gene expression as adapter molecules 'that translate codons in mRNA into amino acids in protein. Since their discovery in the 1950s, extensive studies have clearly defined their biological properties [7]. tRNAs are 70-90 nt in length and form as cloverleaf secondary structure containing three major loops (D-, T-, and anticodon loops) and four stems (acceptor-, D-, T-, and anticodon stems) (Figure 1). These loops and stems fold into an L-shaped tertiary structure. Over 500 tRNA genes are encoded in the human genome [8], and tRNAs have long half-lives, estimated on the order of days in tissues[9, 10]. Active transcription from multiple sites and high stability place tRNAs among the most abundant RNA molecules, occupying around 15% of the cellular RNA transcriptome.
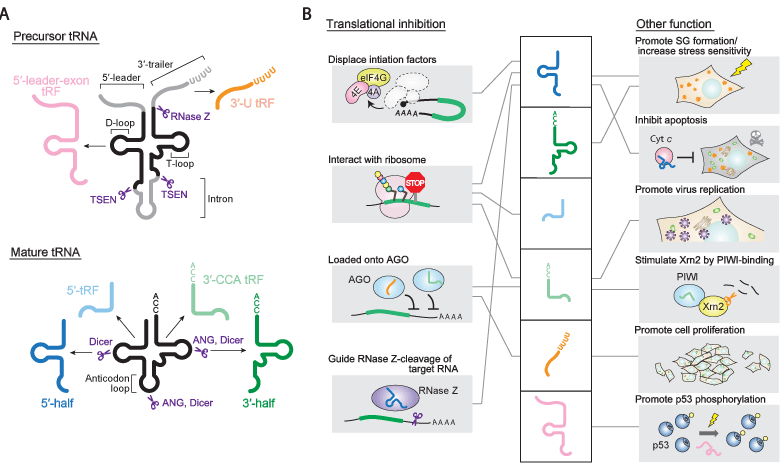
Figure 1: (A) tRNA fragments derived from precursor or mature tRNAs and their processing enzymes. (B) Reported molecular functions of tRNA fragments.
Considering their abundance and well-defined biological role in translation, it is not surprising that RNA fragments from tRNAs were regarded as non-functional degradation intermediates for a long time. The apparent presence of abundant tRNA fragments in early NGS studies was often ignored. However, the combined biochemical evidence from many years of tRNA biology has recently brought the field to a previously unexpected conceptual consensus: specific tRNA fragments are widely expressed not as random degradation products but as functional molecules in many different cells of various organisms [11-15]. The expression of tRNA fragments does not usually affect mature tRNA pools. Instead, it is involved in normal biological processes beyond translation and in diseases; thus, studies of tRNA fragment have provided new insights into tRNA biology. The first evidence of the presence of functional tRNA fragments was reported in 1999; conditioned medium from human urinary bladder carcinoma cells contained tRNA fragments that exhibited an inhibitory effect on endothelial cell growth [16]. In this review, we will summarize the subsequent accumulation of findings, as displayed in Table 1, concerning functional tRNA fragments whose molecular functions and/or associated proteins have been experimentally examined and validated.
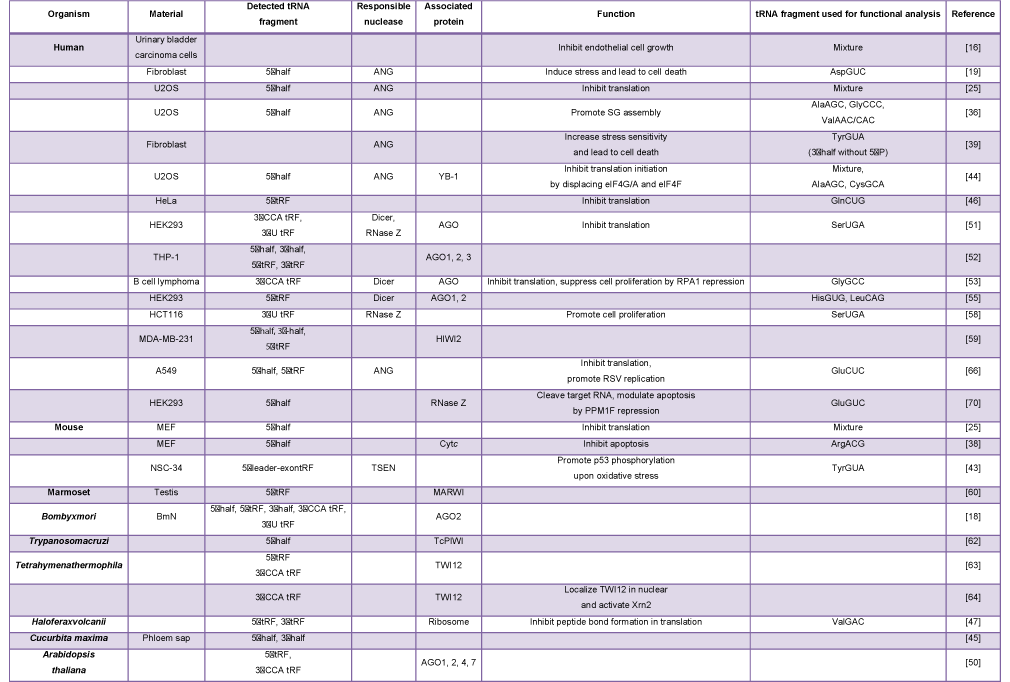
Table 1: Summary of the tRNA fragment studies that investigated molecular functions and/or associated proteins of tRNA fragments.
Classification of tRNA Fragments: tRNA Halves and tRFs
Mature tRNAs are produced from precursor tRNAs (pre-tRNAs), which undergo several steps during maturation [11]. In the first step, pre-tRNAs are transcribed from tRNA genes by RNA polymerase III. The 5'-leader and 3'-trailer sequences of the pre-tRNAs are subsequently removed by endonucleolytic cleavage catalyzed by RNase P and RNase Z, respectively. The trinucleotides "CCA" are then attached to the 3'-termini of tRNAs by CCA-adding enzyme. tRNAs also undergo modification events to create many different non-canonical bases at various positions. Finally, the resultants mature tRNAs are aminoacylated by their cognate aminoacyl-tRNA synthetases, and participate in translation on the ribosome. Both pretRNAs and mature tRNAs serve as substrates for the production of tRNA fragments.
Following the proposed nomenclature [14], tRNA fragments identified so far can be classified into two groups: tRNA halves and tRNA-derived fragments (tRFs)[11-15]. tRNA halves are composed of 30-35 nt fragments derived from either the 5'- or 3'-part of fully matured tRNAs with processed 5'- and 3'-CCA termini (Figure 1). Shorter than tRNA halves, tRFs range from 13-20 nt in length, and can presently be sub classified into four subgroups: 5'-tRFs, 3'-CCA tRFs, 3'-U tRFs, and 5'-leader-exon tRFs (Figure 1). The 5'-tRFs and 3'-CCA tRFs correspond to the 5'- and 3'-parts of mature tRNAs containing processed 5'- and 3'-CCA termini, respectively. Since 5'-leader sequences are absent, 5'-tRFs are formed by cleavage in the D-loop after RNase P-catalyzed removal of 5'-leader sequences. The presence of CCA sequences in 3'-CCA tRFs indicates that the cleavage in the T-loop that produces the 3'-CCA tRFs occurs after 3'-terminal maturation by CCA-adding enzyme. Because these tRFs and tRNA halves coexist (e.g., 5'-tRFs were purified together with 5'- tRNA halves in Dicer-immunoprecipitates) [17-21], the tRNA halves, as well as mature tRNAs, may also be a direct precursor of these tRFs. The 3'-U tRFs are derived from the 3'-leader sequence of pretRNAs that harbor 3'-terminal uridine stretches, which result from transcriptional termination by RNA polymerase III at thymidine tracts. The 5'-leader-exon tRFs, another class of precursor-derived tRFs, include the 5'-leader and the 5'-part of mature tRNAs.
tRNA Halves as Stress-responsive Elements
Eukaryotic tRNA halves were first identified in Tetrahymena as starvation-induced molecules [22]. Their expression was subsequently shown to be triggered in a wide variety of organisms by a variety of stress stimuli, such as oxidative stress, heat/cold shocks, and UV irradiation [21, 23-30]. Therefore, tRNA halves are also known as tRNA-derived stress-induced RNAs (tiRNAs) [25], although they are also detected under non-stressed conditions [17, 31-33]. In mammalian cells, Angiogenin (ANG), a member of the RNase A super family, was discovered to be the enzyme that cleaves the anticodon loops of mature tRNAs to produce tRNA halves [25, 26]. Rny1p, a member of the RNase T2 family, is responsible for this anticodon cleavage in yeast [24]. RNH1, an ANG inhibitor interacting with ANG in the cytoplasm, was shown to be a regulatory factor for ANG cleavage[25]. Interestingly, a tRNA modification and the enzymes that catalyze it were also shown to be regulatory factors for the production of tRNA halves. A number of tRNAs can be methylated by Dnmt2 and NSun2 methyltransferases; this modification protects the modified tRNAs from stress-induced cleavage [19, 34, 35]. It is noteworthy that the stabilities and abundances of the two fragments supposedly produced from the same anticodon cleavage, the 5'- and 3'-tRNA halves, can be asymmetric depending on the organism and the conditions[19, 21, 26, 31].
Fascinatingly, stress-induced tRNA halves promote the phosphor-eIF2a-independent formation of stress granules (SGs) [36]. Because an SG encompasses stalled translation pre-initiation complexes together with poly A-tailed mRNAs for translational repression and future translational induction [37], tRNA halves may play an important signaling role in regulating gene expression by targeting translation initiation complex. The ability to promote SG assembly varies depending on the species of the tRNA halves. Only 5'-tRNA halves, not 3'-tRNA halves, show SG formation activity; a 5'-tRNA half from tRNAAla shows the strongest activity [36]. These observations raise the possibility that tRNA halves might interact with specific factors through the certain sequence motifs within tRNA. This implies that the generation of tRNA halves may be controlled in both qualitative and quantitative ways to produce different amounts of selected tRNA half-species for adaptation against different stresses.
Recent studies provide compelling evidence that the enhanced expression of tRNA halves is involved in human disease as a source of stress response molecules. NSun2, an RNA methyltransferase whose genetic mutations are associated with neurodevelopmental disorders, methylates a majority of expressed tRNAs to generate the m5C modification [19]. tRNAs lacking this modification in NSun2- mutated patient fibroblasts or NSun2-deficient mice are more susceptible to anticodon cleavage by ANG. As a result, 5'-tRNA halves accumulate in NSun2-deficient cells, which is both necessary and sufficient to trigger cellular stress responses in those cells. Because cellular stress responses often precede apoptosis, it is not surprising that stress responses activated by the accumulation of the tRNA halves result in increased apoptosis in the neurons of NSun2- deficient mice [19]. Contrary to their causative effect on apoptosis, tRNA halves produced from ANG cleavage have also been reported to inhibit apoptosis by interacting with cytochrome c in osmoticallystressed mouse embryonic fibroblasts [38].
The other example of disease-related tRNA fragments has been observed in cells with defective CLP1, which is a multifunctional kinase whose genetic mutations are found in neurodegenerative diseases [39- 41]. CLP1 associates with the tRNA-splicing endonuclease (TSEN) complex and contributes to tRNA splicing by phosphorylating the 5'- ends of 3'-tRNA halves, which are themselves the products of TSEN complex-catalyzed removal of the anticodon intron of pre-tRNAs[42]. CLP1-deficient mice experience neurodegeneration and accumulate a 5'-leader-exon tRF derived from tRNATyr that leads to oxidative stress-induced cell death by promoting p53 phosphorylation [40, 43]. The catalytically-inactive CLP1 mutant derived from patients with neurological diseases impairs TSEN-cleavage of pre-tRNA and increases the sensitivity to oxidative stress [39, 40]. Shaffer et al. further demonstrated the toxicity of the 5'-phosphate-lacking 3'- tRNA half from tRNATyr in fibroblasts and neural cells [39]. Taken together, these results suggest that accumulation of aberrant tRNA fragments resulting from deficient CLP1 and TSEN activities could be involved in the pathogenesis of neurological diseases.
Global Translational Inhibition Induced by tRNA Halves and tRFs
When ANG has been reported to be responsible for the production of tRNA halves under stress conditions, transfection of 5'-tRNA halves, but not 3'-tRNA halves, has been shown to inhibit global translation in human cells [25]. Ivanov et al. then revealed the molecular mechanism of the translational inhibition: a 5'-tRNA half from tRNAAla displaces eIF4G/A and eIF4E/G/A (eIF4F) from uncapped RNA and m7G-capped RNA, respectively [44]. YB-1, a multifunctional DNA/RNA-binding protein, was found to strongly associate with the 5'-tRNA half and mediate the translational inhibition and SG formation. Interestingly, a terminal oligo-G motif present in certain 5'-tRNA halves, such as the 5'-tRNA halves from tRNAAla and tRNACys , has been shown to be required for the translational repression [44]. Because the tRNA halves detected in the cells are not limited to those that contain the motif, it remains to be determined whether or not tRNA halves utilize other mechanisms for translational regulation. Translational inhibition induced by tRNA halves were also suggested in plant. The phloem sap of pumpkin contains many 5'- and 3'-tRNA halves, and sap-derived RNA exhibits global translational inhibition [45]. The translational inhibition is likely to be mediated by tRNA halves (or nicked-tRNAs), because the study also showed that RNase-treated, fragmented tRNAs inhibited translation.
In addition to tRNA halves, tRFs have also been reported to be involved in global translational inhibition in human cells. A 19 nt 5'- tRF derived from tRNAGln decreased the expression of a reporter gene that did not have a sequence complementary to that of the 5'-tRF, suggesting that non-specific translational repression is mediated by this 5'-tRF [46]. It is intriguing that the conserved "GG" dinucleotide in the 3'-region of this tRF is required for translation repression. Because 5'-tRFs were detected in the polysome fraction, interaction of 5'-tRFs with the ribosome may contribute to translation repression [46]. Indeed, a 5'-tRF derived from tRNAVal , one of the most abundant tRFs in halophilic archaea, associates with the ribosome and represses translation by interfering with peptide bond formation [47].
tRFs Bound to Argonaute Family Proteins
Although the above-mentioned translational inhibitions are not sequence-specific and their mechanisms are completely different from that of miRNA-mediated regulation, it has become apparent that some tRNA fragments act as miRNAs or other small regulatory RNAs by binding to Argonaute family proteins. Argonaute family proteins can be divided into two sub-clades: AGO and PIWI [2-4]. AGO proteins are ubiquitously expressed in all tissues and bind to miRNAs and siRNAs that are 20-23 nt in length. In contrast, PIWI proteins are predominantly expressed in the germ line (and sometimes in cancers [48]), and interact with 26-31 nt PIWIinteracting RNAs (piRNAs). miRNAs and siRNAs are processed from hairpin-structured or double-stranded precursor RNAs by Dicer endonuclease, while piRNAs are believed to be generated from long single-stranded RNAs by Dicer-independent biogenesis.
The development of antibodies against Argonaute family proteins has enabled the Immunoprecipitation of small RNA fractions associated with these endogenous proteins. This has allowed identification of many tRNA fragments, as well as the abovementioned small regulatory RNAs, as Argonaute-binding small RNAs. Immunoprecipitation of AGO proteins identified significant amounts of tRNA fragments in Drosophila [49], Bombyx [18], and Arabidopsis [50]. In human, 5'-tRFs, 3'-CCA tRFs, and 3'-U tRFs have been reported to bind to AGO proteins [51-54]. Although mature tRNAs do not meet the structural criteria for canonical Dicer substrates, the biogenesis of some tRFs have been reported to be dependent on Dicer in human [51, 53, 55, 56] and mouse [57]. Alternatively, ANG or other enzymes were proposed to be responsible for Dicer-independent biogenesis of AGO-bound tRFs [54]. RNaseZ cleavage produces 3'-U tRFs from pre-tRNAs in a Dicer-independent manner [51, 58]. Thus, tRFs produced through both Dicer-dependent and-independent path ways have been reported to bind to AGO proteins. It is noteworthy that tRFs are asymmetrically loaded onto respective AGO proteins [51, 52, 54]. For example, among the four human AGO proteins (AGO1-4), 3'-CCA tRFs and 3'-U tRFs were shown to preferably associate with AGO3 and AGO4 compared to the other two [51]. This asymmetric loading of tRFs onto different AGO proteins could have significance when considering their function and biogenesis pathway.
Immunoprecipitation of PIWI proteins has also led to the identification of tRNA fragments in human [59], marmoset [60], Drosophila [61], Trypanosoma [62], and Tetrahymena [63, 64], suggesting that tRNA fragments are widely conserved PIWI-binding factors. The biogenesis mechanisms leading to these piRNAs-like tRNA fragments remain to be determined. A PIWI protein essential for growth in Tetrahymena, TWI12, binds tightly to 3'-CCA tRFs [63]. TWI12 itself is proposed to recognize the tertiary structure of full-length tRNAs and cleave them within the T-loop to generate 3'-CCA tRFs. The production of 3'-CCA tRFs and their interaction with TWI12 are necessary to stabilize, localize, and stimulate Xrn2 exonuclease for nuclear RNA processing, indicating a novel role for tRFs in RNA metabolism [64].
Targeted Gene Silencing by tRFs and tRNA Halves
When tRFs are bound to AGO proteins, they are expected to act as miRNAs and repress specific gene expression by recognizing complementary target mRNAs [2-4]. Therefore, targets for these tRFs can be explored through bioinformatics approaches or through biochemical purification of AGO-bound target RNAs. As in the case of miRNAs [2-4], imperfect tRF-target RNA base pairing could induce translational silencing, whereas perfect base pairing could trigger exonucleolytic decay of the target mRNAs.
One of the first studies describing the function of AGO-tRF complexes was obtained from virus research. It has been long known that retroviruses use host tRNA as a primer for reverse transcription during the first step of retroviral replication. Yeng et al. showed that an 18 nt 3'-CCA tRF derived from one such tRNAs, tRNALys , associates with AGO2 and targets the primer-binding site (PBS) of human immunodeficiency virus type 1 (HIV-1) [65]. This 3'-CCA tRF directs AGO2-mediated cleavage of the complementary PBS sequence, thereby silencing PBS-containing reporter gene and an HIV-1 gene. Because endogenous retroviral sequences are found extensively in the human genome and 3'-tRFs are highly complementary to them [54], tRF-directed pathways may also have a role in silencing endogenous viral elements. While these reports suggest tRFs act as negative factors for viral expression, positive effects of tRFs on viral replication have also been reported. Infection with human respiratory syncytial virus (RSV) leads to accumulation of 5'-tRFs that are generated by ANGmediated cleavage at sites adjacent to 5'-end of the anticodon-loop [66]. At least one of the accumulated 5'-tRFs, a 5'-tRF derived from tRNAGlu, has been shown to repress reporter gene expression and promote RSV replication. As another example of a tRF with a positive effect, a 3'-CCA tRF from tRNAPro has been suggested to function as a primer for reverse transcription of human T-cell leukemia virus-1(HTLV-1) [67].
Maute et al. also reported an example of a tRF that functions as a miRNA in mature human B cells [53]. In these cells, a 22nt 3'-CCA tRF derived from tRNAGly is generated in a Dicer-dependent manner and associated with AGO proteins. The 3'-CCA tRF was shown to repress the expression of target mRNAs in a sequence-specific manner. RPA1, an essential gene for DNA dynamics and repair, was identified as the endogenous target containing complementary sequences in its 3'-UTR. Indeed, expression of this tRF suppresses cell proliferation and modulates the response to DNA damage, indicating the clear biological importance of this tRF [53]. While the endogenous targets and biological functions of most of AGO-binding tRFs remain obscure, Haussecker et al. used reporter assay to confirm that AGObound 3'-CCA tRFs and 3'-U tRFs silenced complementary target RNAs [51]. One of the tRFs used in their analysis, a 3'-U tRF derived from tRNASerUGA, was also reported to promote cell proliferation by Lee et al.[58], which validates its cellular role, although it is unknown whether or not interaction with AGO is required for the function of this tRF. Accumulation of further evidence of the functionality and endogenous biological roles of AGO-bound tRFs could result in clearcut designation of these tRFs as small regulatory RNA molecules.
In addition to the AGO-bound tRFs, tRNA fragments are implicated in the AGO-independent sequence-specific silencing of target RNAs by binding to tRNA processing enzymes. Nashimoto et al. showed that RNase Z, when associated with a small guide RNA (sgRNA), can cleave target mRNAs bearing a binding site complementary to the sequence of sgRNAs [68, 69]. Intriguingly, this silencing, referred to as TRUE (tRNase ZL-utilizing efficacious) silencing, can utilize 5'-tRNA halves as sgRNAs in vitro and in vivo for efficient silencing of complementary target RNAs [70]. PPM1F mRNA was identified as one of the endogenous targets of a 5'-tRNA half derived from tRNAGln. These findings imply a physiological role for the complex of RNase Z and 5'-tRNA halves in apoptosis, because both RNase Z and PPM1F are implicated in the regulation of apoptosis.
Future Perspectives
An increasing number of reports have revealed the abundant expression of functional tRNA fragments; thus, it now appears highly plausible that cells use tRNAs as a source for small functional RNAs to modulate biological processes beyond translation. However, research on tRNA fragments is still in the beginning stage; information regarding tRNA fragment expression profiles is still fragmented, and the molecular basis behind the biogenesis and function of tRNA fragments remain elusive.
The immediate focus should be to capture the comprehensive repertoire of tRNA fragments in different tissues and cells by creating libraries and accurately profiling them or by re-evaluating available NGS data sets. However, the biological properties of tRNA fragments could make these attempts challenging. One such property is the presence of post-transcriptional modifications with in tRNA fragments. Because tRNA fragments contain noncanonical modified nucleotides of mature tRNAs, many of which inhibit Watson-Crick base paring and thus cause arrest of reversetranscription or abnormal read through by reverse transcriptase [71], using tRNA fragments to prepare cDNA could result in mutations or severe reductions in the quantity of the resulting cDNAs. Therefore, tRNA fragments from heavily modified mature tRNAs could be underrepresented in sequencing data. This problem is inevitable with any sequencing technology used for detection and quantification of RNA, because there is no experimental methodology to remove all tRNA modifications. Another property that must be considered is the terminal modification of tRNA fragments. The tRNA fragments derived from the 3'-part of mature tRNAs, such as 3'-tRNA halves and 3'-CCA tRFs, contain an amino acid at their 3'-end. Furthermore, the 5'-part of ANG-generating fragments such as 5'-tRNA halves could contain a cyclic-phosphate at their 3'-end [72]. The preparation of cDNA without adequate procedures to remove these terminal modifications will result in severe under representation of these tRNA fragments. If tRNA fragments are produced from the 5'-leader sequences of pre-tRNAs, their 5'-end would contain a tri phosphate modification [43]. Normal RNA sequencing methods do not include a procedure to remove 5'-triphosphates, which may be one of the reasons why tRFs from 5'-leader sequences have not yet been widely discovered. It is crucial to produce libraries and interpret sequencing data from these biochemical perspectives and to confirm the observations using less-biased alternative techniques, such as northern blot or tRNA microarray [28, 73].
To discriminate functional tRNA fragments from "meaningless" degradation products, it is imperative to biochemically elucidate the factors in their biogenesis and to determine their molecular functions. This could be achieved by analyzing their localization, identifying the proteins with which they interact, and examining various biological phenomena in their presence and absence. Regarding localization, for example, 3'-U tRFs accumulate in the cytosol, although their biogenesis occurs in the nucleus [74], suggesting that 3'-UtRFsare actively exported and exert their function in the cytoplasm. As AGO-binding property of RNAs speculates miRNA-like role for the RNAs, identification of interacting proteins is particularly important to address the biological role of tRNA fragments. Bioinformatics studies with support from biochemical assays have identified numerous novel tRNA-interacting proteins [75], implying that the biological functions of tRNAs and their fragments may be way beyond our expectations. Further combination of computational and biochemical efforts will significantly advance our understanding of functional tRNA fragments and expand our knowledge regarding the tRNA world.
Acknowledgments
We are grateful to the members of the authors' lab for helpful discussions. Work in the lab on this topic was supported in part by a grant (GM106047 to YK) from the National Institutes of Health.
References
- Esteller M . Non-coding RNAs in human disease. See comment in PubMed Commons below Nat Rev Genet. 2011; 12: 861-874.
- Farazi TA1, Juranek SA, Tuschl T . The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. See comment in PubMed Commons below Development. 2008; 135: 1201-1214.
- Ghildiyal M1, Zamore PD . Small silencing RNAs: an expanding universe. See comment in PubMed Commons below Nat Rev Genet. 2009; 10: 94-108.
- Kim VN1, Han J, Siomi MC . Biogenesis of small RNAs in animals. See comment in PubMed Commons below Nat Rev Mol Cell Biol. 2009; 10: 126-139.
- Kozomara A1, Griffiths-Jones S . miRBase: annotating high confidence microRNAs using deep sequencing data. See comment in PubMed Commons below Nucleic Acids Res. 2014; 42: D68-73.
- Friedman RC1, Farh KK, Burge CB, Bartel DP . Most mammalian mRNAs are conserved targets of microRNAs. See comment in PubMed Commons below Genome Res. 2009; 19: 92-105.
- RajBhandary, U.L. and D. Soll, Transfer RNA in Its Fourth Decade, in tRNA: Structure, Biosynthesis and Function., D. Soll and U.L. RajBhandary, Editors. 1995, American Society for Microbiology: Washington, D.C. p. 1-4.
- Lowe TM1, Eddy SR . tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. See comment in PubMed Commons below Nucleic Acids Res. 1997; 25: 955-964.
- Nwagwu M, Nana M . Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. See comment in PubMed Commons below J Embryol Exp Morphol. 1980; 56: 253-267.
- Kanerva, P.A. and P.H. Maenpaa, Codon-specific serine transfer ribonucleic acid degradation in avian liver during vitellogenin induction. Acta Chem Scand B, 1981. 35: 379-385.
- Raina M, Ibba M . tRNAs as regulators of biological processes. See comment in PubMed Commons below Front Genet. 2014; 5: 171.
- Gebetsberger J1, Polacek N2 . Slicing tRNAs to boost functional ncRNA diversity. See comment in PubMed Commons below RNA Biol. 2013; 10: 1798-1806.
- Garcia-Silva MR1, Cabrera-Cabrera F2, Güida MC3, Cayota A4 . Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. See comment in PubMed Commons below Genes (Basel). 2012; 3: 603-614.
- Sobala A1, Hutvagner G . Transfer RNA-derived fragments: origins, processing, and functions. See comment in PubMed Commons below Wiley Interdiscip Rev RNA. 2011; 2: 853-862.
- Phizicky EM1, Hopper AK . tRNA biology charges to the front. See comment in PubMed Commons below Genes Dev. 2010; 24: 1832-1860.
- Zhao H1, Bojanowski K, Ingber DE, Panigrahy D, Pepper MS, Montesano R, Shing Y . New role for tRNA and its fragment purified from human urinary bladder carcinoma conditioned medium: inhibition of endothelial cell growth. See comment in PubMed Commons below J Cell Biochem. 1999; 76: 109-117.
- Nowacka M1, Strozycki PM, Jackowiak P, Hojka-Osinska A, Szymanski M, Figlerowicz M . Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. See comment in PubMed Commons below Plant Mol Biol. 2013; 83: 191-204.
- Nie Z1, Zhou F, Li D, Lv Z, Chen J, Liu Y, Shu J . RIP-seq of BmAgo2-associated small RNAs reveal various types of small non-coding RNAs in the silkworm, Bombyx mori. See comment in PubMed Commons below BMC Genomics. 2013; 14: 661.
- Blanco S1, Dietmann S1, Flores JV1, Hussain S1, Kutter C2, Humphreys P1, Lukk M2 . Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. See comment in PubMed Commons below EMBO J. 2014; 33: 2020-2039.
- Durdevic Z1, Mobin MB, Hanna K, Lyko F, Schaefer M . The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. See comment in PubMed Commons below Cell Rep. 2013; 4: 931-937.
- Hsieh LC1, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH . Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. See comment in PubMed Commons below Plant Physiol. 2009; 151: 2120-2132.
- Lee SR1, Collins K . Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. See comment in PubMed Commons below J Biol Chem. 2005; 280: 42744-42749.
- Thompson DM1, Lu C, Green PJ, Parker R . tRNA cleavage is a conserved response to oxidative stress in eukaryotes. See comment in PubMed Commons below RNA. 2008; 14: 2095-2103.
- Thompson DM1, Parker R . The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. See comment in PubMed Commons below J Cell Biol. 2009; 185: 43-50.
- Yamasaki S1, Ivanov P, Hu GF, Anderson P . Angiogenin cleaves tRNA and promotes stress-induced translational repression. See comment in PubMed Commons below J Cell Biol. 2009; 185: 35-42.
- Fu H1, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R . Stress induces tRNA cleavage by angiogenin in mammalian cells. See comment in PubMed Commons below FEBS Lett. 2009; 583: 437-442.
- Thompson DM1, Parker R . Stressing out over tRNA cleavage. See comment in PubMed Commons below Cell. 2009; 138: 215-219.
- Saikia M1, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P . Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. See comment in PubMed Commons below J Biol Chem. 2012; 287: 42708-42725.
- Li Y1, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH . Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. See comment in PubMed Commons below Nucleic Acids Res. 2008; 36: 6048-6055.
- Garcia-Silva MR1, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C . A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. See comment in PubMed Commons below Mol Biochem Parasitol. 2010; 171: 64-73.
- Dhahbi JM1, Spindler SR, Atamna H, Yamakawa A, Boffelli D, Mote P, Martin DI . 5' tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. See comment in PubMed Commons below BMC Genomics. 2013; 14: 298.
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L . A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. See comment in PubMed Commons below Cell Res. 2012; 22: 1609-1612.
- Galizi R1, Spano F, Giubilei MA, Capuccini B, Magini A, Urbanelli L, Ogawa T . Evidence of tRNA cleavage in apicomplexan parasites: Half-tRNAs as new potential regulatory molecules of Toxoplasma gondii and Plasmodium berghei. See comment in PubMed Commons below Mol Biochem Parasitol. 2013; 188: 99-108.
- Goll MG1, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG . Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. See comment in PubMed Commons below Science. 2006; 311: 395-398.
- Schaefer M1, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F . RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. See comment in PubMed Commons below Genes Dev. 2010; 24: 1590-1595.
- Emara MM1, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF . Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. See comment in PubMed Commons below J Biol Chem. 2010; 285: 10959-10968.
- Adjibade P1, Mazroui R2 . Control of mRNA turnover: Implication of cytoplasmic RNA granules. See comment in PubMed Commons below Semin Cell Dev Biol. 2014; 34C: 15-23.
- Saikia M1, Jobava R2, Parisien M3, Putnam A4, Krokowski D1, Gao XH1, Guan BJ1 . Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. See comment in PubMed Commons below Mol Cell Biol. 2014; 34: 2450-2463.
- Schaffer AE1, Eggens VR2, Caglayan AO3, Reuter MS4, Scott E1, Coufal NG1, Silhavy JL1 . CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. See comment in PubMed Commons below Cell. 2014; 157: 651-663.
- Karaca E1, Weitzer S2, Pehlivan D1, Shiraishi H2, Gogakos T3, Hanada T4, Jhangiani SN5 . Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. See comment in PubMed Commons below Cell. 2014; 157: 636-650.
- Weitzer S1, Hanada T, Penninger JM, Martinez J . CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. See comment in PubMed Commons below Wiley Interdiscip Rev RNA. 2014; .
- Weitzer S1, Martinez J . The human RNA kinase hClp1 is active on 3' transfer RNA exons and short interfering RNAs. See comment in PubMed Commons below Nature. 2007; 447: 222-226.
- Hanada T1, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R . CLP1 links tRNA metabolism to progressive motor-neuron loss. See comment in PubMed Commons below Nature. 2013; 495: 474-480.
- Ivanov P1, Emara MM, Villen J, Gygi SP, Anderson P . Angiogenin-induced tRNA fragments inhibit translation initiation. See comment in PubMed Commons below Mol Cell. 2011; 43: 613-623.
- Zhang S1, Sun L, Kragler F . The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. See comment in PubMed Commons below Plant Physiol. 2009; 150: 378-387.
- Sobala A1, Hutvagner G . Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. See comment in PubMed Commons below RNA Biol. 2013; 10: 553-563.
- Gebetsberger J1, Zywicki M, Künzi A, Polacek N . tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. See comment in PubMed Commons below Archaea. 2012; 2012: 260909.
- Suzuki R1, Honda S, Kirino Y . PIWI Expression and Function in Cancer. See comment in PubMed Commons below Front Genet. 2012; 3: 204.
- Kawamura Y1, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN . Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. See comment in PubMed Commons below Nature. 2008; 453: 793-797.
- Loss-Morais G1, Waterhouse PM, Margis R . Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. See comment in PubMed Commons below Biol Direct. 2013; 8: 6.
- Haussecker D1, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA . Human tRNA-derived small RNAs in the global regulation of RNA silencing. See comment in PubMed Commons below RNA. 2010; 16: 673-695.
- Burroughs AM1, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO . Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. See comment in PubMed Commons below RNA Biol. 2011; 8: 158-177.
- Maute RL1, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R . tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. See comment in PubMed Commons below Proc Natl Acad Sci U S A. 2013; 110: 1404-1409.
- Li Z1, Ender C, Meister G, Moore PS, Chang Y, John B . Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. See comment in PubMed Commons below Nucleic Acids Res. 2012; 40: 6787-6799.
- Cole C1, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ . Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. See comment in PubMed Commons below RNA. 2009; 15: 2147-2160.
- Morin RD1, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y . Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. See comment in PubMed Commons below Genome Res. 2008; 18: 610-621.
- Babiarz JE1, Ruby JG, Wang Y, Bartel DP, Blelloch R . Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. See comment in PubMed Commons below Genes Dev. 2008; 22: 2773-2785.
- Lee YS1, Shibata Y, Malhotra A, Dutta A . A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). See comment in PubMed Commons below Genes Dev. 2009; 23: 2639-2649.
- Keam SP1, Young PE2, McCorkindale AL2, Dang TH2, Clancy JL2, Humphreys DT2, Preiss T2 . The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. See comment in PubMed Commons below Nucleic Acids Res. 2014; 42: 8984-8995.
- Hirano T1, Iwasaki YW1, Lin ZY2, Imamura M3, Seki NM4, Sasaki E5, Saito K1 . Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. See comment in PubMed Commons below RNA. 2014; 20: 1223-1237.
- Brennecke J1, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ . Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. See comment in PubMed Commons below Cell. 2007; 128: 1089-1103.
- Garcia-Silva, M.R., et al., A particular set of small non-coding RNAs is bound to the distinctive Argonaute protein of Trypanosoma cruzi: insights from RNA-interference deficient organisms. Gene, 2014. 538: 379-384.
- Couvillion MT1, Sachidanandam R, Collins K . A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. See comment in PubMed Commons below Genes Dev. 2010; 24: 2742-2747.
- Couvillion MT1, Bounova G, Purdom E, Speed TP, Collins K . A Tetrahymena Piwi bound to mature tRNA 3' fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. See comment in PubMed Commons below Mol Cell. 2012; 48: 509-520.
- Yeung ML1, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT . Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. See comment in PubMed Commons below Nucleic Acids Res. 2009; 37: 6575-6586.
- Wang Q1, Lee I, Ren J, Ajay SS, Lee YS, Bao X . Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. See comment in PubMed Commons below Mol Ther. 2013; 21: 368-379.
- Ruggero K1, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A . Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. See comment in PubMed Commons below J Virol. 2014; 88: 3612-3622.
- Nashimoto M . Specific cleavage of target RNAs from HIV-1 with 5' half tRNA by mammalian tRNA 3' processing endoribonuclease. See comment in PubMed Commons below RNA. 1996; 2: 523-524.
- Nakashima A1, Takaku H, Shibata HS, Negishi Y, Takagi M, Tamura M, Nashimoto M . Gene silencing by the tRNA maturase tRNase ZL under the direction of small-guide RNA. See comment in PubMed Commons below Gene Ther. 2007; 14: 78-85.
- Elbarbary RA1, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H . Modulation of gene expression by human cytosolic tRNase Z(L) through 5'-half-tRNA. See comment in PubMed Commons below PLoS One. 2009; 4: e5908.
- Kellner S1, Burhenne J, Helm M . Detection of RNA modifications. See comment in PubMed Commons below RNA Biol. 2010; 7: 237-247.
- Rybak SM1, Vallee BL . Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. See comment in PubMed Commons below Biochemistry. 1988; 27: 2288-2294.
- Dittmar KA1, Goodenbour JM, Pan T . Tissue-specific differences in human transfer RNA expression. See comment in PubMed Commons below PLoS Genet. 2006; 2: e221.
- Liao JY1, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ . Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3' trailers. See comment in PubMed Commons below PLoS One. 2010; 5: e10563.
- Parisien M1, Wang X, Perdrizet G 2nd, Lamphear C, Fierke CA, Maheshwari KC, Wilde MJ . Discovering RNA-protein interactome by using chemical context profiling of the RNA-protein interface. See comment in PubMed Commons below Cell Rep. 2013; 3: 1703-1713.