
Special Aricle - Protein Synthesis
J Mol Biol & Mol Imaging. 2015;2(2): 1018.
The Quality is First: Tagging Nascent Aberrant Proteins for Destruction
Lorena Gonzalez López and Fidel de la Cruz Hernández-Hernández*
Infectomics and Molecular Pathogenesis Department, CINVESTAV-IPN, México
*Corresponding author: Hernández-Hernández. F.C., Infectomics and Molecular Pathogenesis Department, Center for Research and Advanced Studies of the National Polytechnical Institute (CINVESTAV-IPN), National Polytechnic InstituteAvenue # 2508, Colonia San Pedro Zacatenco, DelegaciónGustavo A. Madero 07360, México D.F., México
Received: May 05, 2015; Accepted: May 14, 2015; Published: May 15, 2015
Keywords
Protein quality control; Ubiquitin; Proteasome; Protein folding.
Abbreviations
ROS: Oxygen Reactive Species; QC: Protein Quality Control; UPS: Ubiquitin Proteasome System; NMD: Nonsense-Mediated Decay; NGD: No-Go Decay; NSD: Non Stop-Decay; mRNA: Messenger RNA; RQC: Ribosome Quality Control; NAC: Nascent Polypeptide Associated Complex; RAC: Ribosome Associated Complex
Editorial
For all the molecular processes that a cell needs to survive, divide or fulfill, billions of proteins have to be produced. To this end, millions of ribosomes are constantly working, with a synthesis rate of approximately six amino acids per second [1]. This makes the protein synthesis a process with a high probability of errors not only regarding the inefficient folding but also errors in translation and the presence of aberrant mRNAs. In addition, nascent proteins are more susceptible to environmental changes such as high temperatures or oxygen reactive species (ROS). Thus, eukaryotic cells have developed protein quality control (QC) pathways to prevent the accumulation of aberrant proteins that cells identify, label and direct to destruction mainly by the ubiquitin-proteasome system (UPS) [2,3]. Ubiquitin is a 76 amino acid molecule that is covalently attached to protein substrates by a sequential enzymatic cascade catalyzed by three classes of enzymes namely E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases. E1 enzymes activate ubiquitin in an ATP-dependent fashion, this enzyme transfer ubiquitin to the E2, which further will be used for ubiquitination of E3 bound substrates [4]. E3 therefore are the enzymes responsible to selectively recognize the proteins to be tagged for degradation.
During translation, ribosomes can stop during elongation and stall for different reasons, mainly due to mRNA defects. The ribosome stalling activates both protein and mRNA QC pathways. To date, there are three QC pathways recognized in eukaryotes: nonsense-mediated decay (NMD), no-go decay (NGD), and nonstop-decay (NSD). The NMD is activated by the translation of mRNAs containing premature termination codons, and the NGD pathway is activated with stable mRNA hairpin structures, rare codons, and positively charged polylysine or polyarginine residues. Finally, the NSD pathway is activated by “non-stop codon” (those lacking of a stop codon) and damaged mRNAs [2,5].
QC pathways have been deeply studied in terms of mRNA turnover; however, the mechanisms for nascent polypeptide degradation or ribosome recycling have been matter of study only in the recent years. The most studied pathways are the NSD and NGD that are activated by a stalled ribosome. In yeasts, the Hbs1-Dom34 complex, two translation elongation factors (homologs of eFR3-eFR1, respectively), binds to the ribosome probably as a heterodimer. The Hbs1-Dom34 complex together with the ribosome recycling ATPase Rli1 promotes disassembly of ribosomes [6,7]. After ribosome splitting, the E3 ligase Ltn1, together with the ribosome quality control 1 (Rqc1), and the translation-associated element 2 (Tae2) are recruited and the aberrant nascent peptide is ubiquitinated [8]. The association of these three proteins and the ubiquitinated protein recruits the AAA-ATPase Cdc48, Ufd1, and Npl4 complex. Based on the known activity of Cdc48, it has been proposed that it might extract the polyubiquitinated peptides from the dissociated 60S and escort them to the proteasome for degradation (Figure 1A) [9,10].
Ltn1 was the first E3 ubiquitin ligase discovered in this RQC complex. Ltn1 mutation has been related to neurodegeneration in mice and has been found related to aberrant peptide synthetized from a non-stop mRNA [11]. Another E3 ubiquitin ligase related to these processes is Not4, a subunit of the Ccr4-Not deadenylase complex. This E3 ubiquitin ligase targets polybasic-containing nascent chains. In this case, Not4 seems to associate with 80S and polyribosomes ubiquitinating aberrant nascent polypeptides by the recruitment of the E2 enzymes Ubc4p and Ubc5p as well as the proteasome [12]. In addition to these E3 ligases, Hel2 has been also described in these QC pathways. Hel2 together with Asc1 participate in the polypeptide quality control in the RQC pathway, but unlike Ltn1, this E3 ligase may be participating in an upstream stage since in some cases (polybasic chains), the initial recognition step appears to require Asc1 and Hel2. These factors may mediate RNA cleavage [10]. Furthermore, other E3 ligases, Upf1 and Ubr1 have been identified. These are also related to this RQC pathway stimulating the degradation of stalled protein chains. However, the exact moment of their participation have not been elucidated [2,7,13].
Other translation errors that conduct to protein ubiquitination and degradation are nascent polypeptides not correctly folded due to mutations, errors during transcription and RNA processing among others [14]. In order to avoid this, cells uses different complexes such as the nascent polypeptide associated complex (NAC), the ribosome associated complex (RAC), Hsp70, and chaperonin that interacts with nascent proteins and facilitates its folding; however, this is not always successful. Thus, misfolded proteins are degraded immediately after their synthesis or when those are being synthetized in a process named co-translational protein degradation. This mechanism has not been completely understood; however, some participating proteins have been identified. In yeasts, the nuclear import factor Srp1, and Sts1 couple proteasomes to nascent peptides emerging from the ribosome for degradation [15]. Also, it was found that the E3 ubiquitin ligases Hel2 and Rkr1 work in parallel with other ubiquitin ligases such as Doa10, Hrd1 and Hul5 to mediate co-translational quality control. In this case, the Cdc48 ATPase appears to function also in delivering the nascent proteins to the proteasome (Figure 1B) [16].
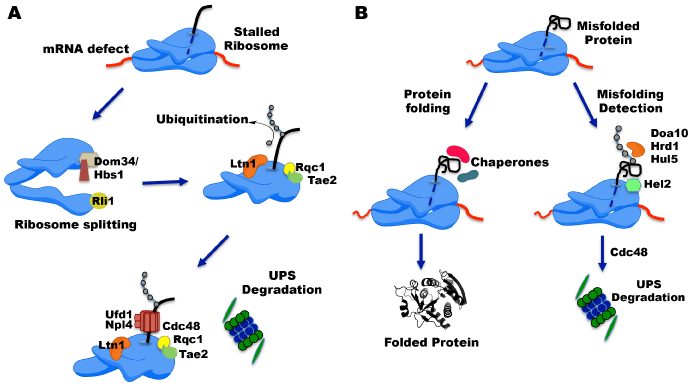
Figure 1: Co-translational ubiquitination of Defective Nascent Proteins. Cotranslational
ubiquitination has been reported that occur in both stalled and
active translation complexes. A) Co-translational ubiquitination in stalled
ribosomes. Stalled ribosomes are dissociated in 60S-petydil-tRNA and 40S
subunits by the Hbs1-Dom34-Rli1 complex. The E3 ligase Ltn1 associates
with the 60S ribosome subunit together with the Ribosome Quality Control
(RQC) complex proteins Rqc1 and Tae2 to add ubiquitin (grey dots) moieties.
After, the Cdc48, Ufd1, and Npl4 complex is recruited and the ubiquitinated
defective nascent protein is degraded by the UPS (Ubiquitin Proteasome
System). B) Co-translational ubiquitination in misfolded nascent proteins. In
general, proteins are efficiently folded with the assistance of chaperones or
the nascent polypeptide associated complex (NAC). When proteins are not
correctly folded E3 ligases, such as Doa10, Hrd1, Hul5, and Hel2, are able
to recognize the misfolded nascent polypeptide and tag them with ubiquitin.
After ubiquitination, the Cdc48 complex is recruited to direct the tagged chain
to the proteasome for its degradation.
In both cases, the molecular mechanisms for aberrant nascent polypeptides are not completely understood. Many questions remain unsolved. How many E3 ligases are working to recognize aberrant nascent proteins? All proteins could be damaged when are synthetized? There are proteins with more potential to be defective? It depends on the translation rate of certain proteins? Which are the signals to recognize a defective/misfolded nascent protein? How does the cell achieve substrate specificity? Which would be the consequences if these processes are not well regulated? Understanding these QC pathways and their mechanisms within the cell is imperative, since some diseases, mainly neurodegenerative, have been related to misfolded protein degradation impairment.
Acknowledgements
FCHH is supported by grants 180514 CONACyT México and CN-10-479 from UC-MEXUS CONACyT. LGL received a Ph.D. scholarship 225016 from CONACyT, México.
References
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147: 789-802.
- Wang F, Canadeo LA, Huibregtse JM. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie. 2015; .
- Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol Cell. 2013; 49: 411-421.
- Heride C, Urbé S, Clague MJ. Ubiquitin code assembly and disassembly. Curr Biol. 2014; 24: R215-220.
- Panasenko OO. The role of the E3 ligase Not4 in cotranslational quality control. Front Genet. 2014; 5: 141.
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010; 330: 369-372.
- Lykke-Andersen J, Bennett EJ. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J Cell Biol. 2014; 204: 467-476.
- Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell. 2014; 55: 880-890.
- Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci U S A. 2013; 110: 5046-5051.
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012; 151: 1042-1054.
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010; 467: 470-473.
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009; 284: 10343-10352.
- Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009; 10: 1265-1271.
- Comyn SA, Chan GT, Mayor T. False start: cotranslational protein ubiquitination and cytosolic protein quality control. J Proteomics. 2014; 100: 92-101.
- Ha SW, Ju D, Xie Y. Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation. The Journal of biological chemistry. 2014; 289: 2701-2710.
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell. 2013; 50: 379-393.