
Case Report
Austin J Obstet Gynecol. 2023; 10(1): 1212.
Genetic Variants and Haplotypes of CDH1 Gene in Polycystic Ovary Syndrome: A Case Control Study in South Indian Women
Bhanoori M1*, Guruvaiah P1, Siddamalla S1, Veena KV1, Govatati S1, Dakshayani L2, Deenadayal M3, and Shivaji S4,5
1Department of Biochemistry, Osmania University, India
2Department of Genetics and Genomics, Yogivemana University, India
3Infertility Institute and Research Centre (IIRC), India
4Centre for Cellular and Molecular Biology (CCMB), India
5Currently at: Brien Holden Eye Research Centre, L V Prasad Eye Institute, India
*Corresponding author: Manjula Bhanoori Department of Biochemistry, Osmania University, Hyderabad – 500 007, India
Received: December 18, 2022; Accepted: January 12, 2022; Published: January 19, 2023
Abstract
Polycystic Ovary Syndrome (PCOS) is a heterogeneous multifactorial endocrine metabolic disorder. The main aim of this study was to investigate the association of Single Nucleotide Polymorphisms (SNPs) of CDH1 gene with the susceptibility to PCOS in South Indian women. This study comprised 105 PCOS cases and 115 controls of South Indian origin. We have genotyped promoter -347G/GA (rs5030625), -160C/A (rs16260) and 3’-UTR +54C/T (rs1801026) of CDH1 gene polymorphisms by PCR-DNA sequencing analysis. The genotype and allele distributions of cases and controls were analysed using Fisher’s exact test. Haplotype frequencies for multiple loci and the standardized disequilibrium coefficient (D’) for pair wise Linkage Disequilibrium (LD) were assessed by Haploview Software. The frequencies of -347 (P=0.02), -160 (P=0.05) and +54(P=0.001) genotypes and G/C/T (P=0.003), G/A/T (P=0.037), GA/A/C (P=0.00008) and GA/A/T (P=0.0008) haplotypes were significantly different between patients and controls. A strong LD was observed between -160 C/A and +54 C/T loci (D’=1), when compared with -347 G/GA and +54 C/T (D’=0.20) or -347 G/GA and -160 C/A (D’=0.07) loci in cases. In conclusion, for the first time we report a significant association of -347 GA, -160 A and +54T 3’-UTR variants in CDH1 gene and PCOS risk in South Indian women.
Keywords: Polycystic ovary syndrome; CDH1 gene polymorphism; PCR-DNA sequencing; Haplotype analysis; South indian women
Abbreviations: CI: Confidence Interval; Χ²: Chi Square; D': Disequilibrium Coefficient; LD: Linkage Disequilibrium; HWE: Hardy– Weinberg Equilibrium; OR: Odds Ratio; PCOS: Polycystic Ovary Syndrome; CDH1: Epithelial Cadherin; CAM: Cell Adhesion Molecules; SNP: Single Nucleotide Polymorphisms; (3’ UTR): 3’ Untranslated Region; MAF: Minor Allelic Frequency
Introduction
Polycystic Ovary Syndrome (PCOS) is the most prevalent endocrine condition in young reproductive aged women. The worldwide prevalence of PCOS is estimated to be 5–10% [14]. PCOS is a multifactorial disorder and is characterized by a combination of clinical (an ovulation and hyperandrogenism), biochemical (excessive and rogen and luteinizing hormone concentrations) and ovarian morphological (polycystic ovaries) features [6]. Fluctuation in hormone levels, stress and obesity are the major cause’s worldwide [1,16]. It can also be associated with type 2 diabetes mellitus (T2DM), psychological effects on quality of life including anxiety and depression, and breast and endometrial cancers. As many as 20% of women with infertility problems (including fecund ability and early pregnancy loss) have been diagnosed with PCOS [5]. The markers of oxidative stress and inflammation are correlated with increased androgen level in women with PCOS. The pathogenesis of this syndrome involves frequent abnormalities on lipid or glucose metabolism [19].
Epithelial Cadherin (CDH1) belongs to Cell Adhesion Molecules (CAM) family, encoded by the CDH1 gene on chromosome 16q22.1. The Single Nucleotide Polymorphisms (SNPs) of the promoter region and 3’-untranslated region (3’-UTR) are critical for transcriptional regulation of CDH1 gene. The most widely studied promoter polymorphisms include -347 G/GA, -160 C/A and 3’-UTR +54 C/T which have a significant effect on CDH1 gene transcription [7]. Available literature shows association of these polymorphisms and risk of developing various human diseases such as Crohn disease, ovarian cancer [9,15] but there are no reports of these SNPs relating to PCOS. We hypothesized that variation in the CDH1 gene may alter the expression of ECadher in which in turn may lead to PCOS susceptibility. In the present case-control study, we determined the distribution of the CDH1-347G/GA (rs5030625), -160C/A (rs16260) and 3’- UTR +54C/T (rs1801026) polymorphisms and their correlation with the risk of developing PCOS in South Indian women.
Materials and Methods
Study Population
One hundred and five (n=105) women of reproductive age 18-40 years (mean age: 27 years) with PCOS and one hundred and ten (n=110) healthy women without PCOS (mean age: 26 years) as controls were recruited at the Infertility Institute and Research Centre (IIRC), Secunderabad, Telangana, India. Blood samples were collected and plasma was removed followed by storage at -20°C until further analysis. Informed written consent was obtained from all subjects prior to participation in this study. The study was approved by ethical committee and review board of Centre for Cellular and Molecular biology (CCMB), Hyderabad. All the participants included in study were of South Indian origin (Dravidian linguistic group).
Cases were selected as per the Rotterdam consensus criteria to diagnose PCOS [20]. All subjects (PCOS cases and controls) were, non-pregnant and non-smokers. Criteria for the diagnosis of PCOS included oligoovulation (cycles longer than 35 days or less than 26 days, elevated free testosterone levels (0.5 ng/dl; the cut-off level for free testosterone level was the mean ±2 SD according to normal levels in controls), oligomenorrhea or amenorrhea. In accordance with the above criteria polycystic ovary morphology was determined by transvaginal ultrasonography, which defines PCOS as the presence of 12 or more small follicles (2 to 9 mm) in each ovary.
Control subjects had no signs of menstrual dysfunction and their glucose tolerance, androgen levels were within normal range, and no family history of hirsutism, type 2 diabetes mellitus, and infertility. The Body Mass Index (BMI) was calculated as body weight (kg) divided by body height squared (m2). The demographic and biochemical characteristics of PCOS women and controls were summarized in (Supplementary Table 1). Women with other causes of hyperandrogenism such as hyperprolactinemia, Cushing syndrome, androgen-secreting tumours and non-classic congenital hyperplasia, were excluded from this study.
S.No.
Variable
PCOS (n =105)
Controls (n =110)
1
Age (years)
27.12±5.05
26.36±4.43
2
Weight (kg)
61.68±9.44
57.68±7.69
3
BMI (kg/m2)
24.29±4.12
23.77±2.92
4
LH (mIU/ml)
8.04±2.15
5.33±1.60
5
FSH (mIU/ml)
4.81±2.21
6.07±1.66
6
LH:FSH
2.44±2.35
0.9±0.21
7
Presence of overweight
17 (16.19%)
36 (32.72%)
8
Presence of obesity
42 (40%)
33 (30%)
Data are given as mean ± S.D or n (%).
Supplementary Table 1: Clinical characteristic features of cases and controls.
Genetic Analysis
Genotyping of CDH1variants-347 G/GA, -160 C/A, 3'-UTR +54 C/T was performed by PCR and sequencing analysis as per the protocols described earlier [11]. PCR was carried out in a total reaction volume of 25 μl, containing 50 ng genomic DNA, 1X Taq polymerase buffer (1.5mM MgCl2), 2–6 p mole of each primer, and 0.25Units of Amplitaq DNA polymerase (Perkin Elmer, Foster City, USA). The primers and PCR conditions were summarized in (Table 1). PCR amplification was performed in a programmable thermal cycler gradient PCR system (Eppendorf AG, Hamburg, Germany). PCR products were analyzed by 1.5% agarose gel stained with ethidium bromide and then sequenced with a Taq-Dye deoxy-terminator cycle sequencing kit (Applied BioSystems, USA) using an automated ABI 3770 DNA sequencer (Applied Bio Systems, USA). Genotype calling was performed by using Chromas V.2 software (Technelysium Ltd., Australia).
Gene
SNP
Primers
PCR conditions
Reference
CDH1
-347 G/GA& -160 C/A
F : 5'-CGCCCCGACTT
GTCTCTCTAC-3'
R : 5'-GGCCACAGCDenaturation: 960C (5 min);
35 cycles : 94°C (40 Sec);
66°C (45 Sec);72°C (50 Sec);Govatati et al., 2012
+54 C/T
CAATCAGCA-3'
F:5'-CAGACAAAGAC
CAGGACTAT-3'
R:5'-AAGGGAGCTGA
AAAACCACCAGC-3'Extension: 72°C(10 min)
Denaturation: 960C (5 min);
35 cycles: 94°C (40 Sec);
56°C (45 Sec);72°C (50 Sec);
Extension: 72°C(10 min)PCR: Polymerase Chain Reaction
Table 1: Primers and PCR conditions used in this study.
Statistical Analysis
Statistical analysis was performed using the SPSS statistical package (V 11.0). Fisher's exact test was used for testing the genotype distribution among subjects for Hardy–Weinberg Equilibrium (HWE) and the allele ratios and genotype distributions of cases and controls. The odds ratio and 95% Confidence Interval (CI) values were calculated using the online Vassar Stats Calculator (http://www.faculty.vassar.edu/lowry/VassarStats. html). Haplotype frequencies for multiple loci and the standardized disequilibrium coefficient (D') for pair-wise Linkage Disequilibrium (LD) were assessed by Haploview Software [4]. The P-value below <0.05 was considered statistically significant.
Results
All subjects (n=215) were successfully genotyped. The genotype distributions of individual SNPs, as well as allele system, were all in Hardy-Weinberg equilibrium (P > 0.05) in both cases and controls.
Genotyping of CDH1 SNPs
-347G/GApolymorphism: The distribution of CDH1 -347 G/GA genotypes in women with PCOS and controls is shown in (Table 2). There was a significant reduction of the genotype (G/G) frequency and elevation of the mutant genotype (GA/GA) frequency in patients as compared to controls, indicating that ‘GA’ allele might confer risk to PCOS and ‘G’ allele may provide protection against development of the disease. Sequence analyses of the 448 bp product of the CDH1 promoter region SNP are shown in (Figure 1(A)).
Genotypes/Alleles
PCOS (%)n = 105
Controls (%)n = 110
Chi-Square
(X2)P-value
Odds
Ratio95% CI
CDH1 (-347 G/GA)
Genotypesa
G/G
48 (45.71)
64 (58.18)
6.079
0.0478
Ref.
G/GA
42 (40.00)
40 (36.36)
0.714
0.4030- 1.2660
GA/GA
15 (14.29)
6 (5.45)
0.300
0.1084 - 0.8303
Allelesb
G
138 (65.71)
168 (76.36)
5.938
0.0148
Ref.
GA
72 (34.29)
52 (23.64)
0.5933
0.3891 - 0.9046
CDH1 (-160 C/A)
Genotypesa
C/C
49 (46.67)
58 (52.73)
5.239
0.0728
Ref.
C/A
39 (37.14)
45 (40.91)
0.975
0.5495 - 1.7292
A/A
17 (16.19)
7 (6.36)
0.357
0.1341 - 0.9501
Allelesb
C
137 (65.24)
161 (73.18)
3.187
0.0742
Ref.
A
73 (34.76)
59 (26.82)
0.688
0.4555 - 1.0382
CDH1 (+54 C/T)
Genotypesa
C/C
51 (48.57)
77 (70.00)
11.589
0.0030
Ref.
C/T
44 (41.90)
30 (27.27)
0.452
0.2519 - 0.8095
T/T
10 (95.23)
3 (2.73)
0.199
0.0521 - 0.7572
Allelesb
C
146 (69.52)
184 (83.64)
11.990
0.0005
Ref.
T
64 (30.48)
36 (16.36)
0.446
0.2810- 0.7088
Ref : Reference ; CI: confidence interval.
aFisher’s exact test (3x2 table at 2 df); P<0.05
bFisher’s exact test (2x2 table at 1 df); P<0.05
Table 2: Risk estimates for the association of CDH1 SNPs in PCOS.
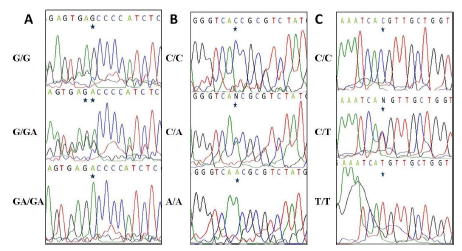
Figure 1: (A) Genotyping of the CDH1 gene -347 G/GA polymorphism
by sequence analysis of the PCR-amplified product using a
forward primer. (B) Genotyping of the CDH1 gene -160 C/A polymorphism
by sequence analysis of the PCR-amplified product using
a forward primer. (C) Genotyping of the CDH1 gene +54 C/T
polymorphism by sequence analysis of the PCR-amplified product
using a forward primer.
-160C/Apolymorphism: The distribution of CDH1 -160 C/A genotypes in women with PCOS and controls is shown in (Table 2). There was a significant reduction of the genotype (C/C) frequency and elevation of the mutant genotype (A/A) frequency in patients as compared to controls indicating that ‘A’ allele might confer risk to PCOS and ‘C’ allele may provide protection against development of the disease. Sequence analyses of the 448 bp product of the CDH1 promoter region SNP are shown in (Figure 1(B)).
+54C/T polymorphism: The distribution of CDH1 +54 C/T genotypes in women with PCOS and controls is shown in (Table 2). There was a significant reduction of the genotype (C/C) frequency and elevation of the mutant genotype (T/T) frequency in patients as compared to controls indicating that ‘T’ allele might confer risk to PCOS and ‘C’ allele may provide protection against development of the disease. Sequence analyses of the 172 bp product of the CDH1 3'-UTR region SNP are shown in (Figure 1(C)).
Haplotype Analysis
To understand the additive effect of studied CDH1 SNPs on the risk of developing PCOS, the haplotype frequencies for multiple loci were calculated (Table 3). The haplotype distributions derived from CDH1 -347 G/GA, -160 C/A and +54 C/T of 3'-UTR polymorphisms significantly differed between the cases and controls (P< 0.05). CDH1 -347/-160/+54 GCC was the most common haplotype in South Indian women. The frequency of haplotypes -347/-160/+54 GCT, GAT, GAAC and GAAT was significantly higher in cases when compared to controls and may significantly increase the risk of developing PCOS. Linkage Disequilibrium (LD) coefficients (D') were calculated to express the strength of linkage between SNPs of the CDH1 gene among cases and controls. A strong linkage disequilibrium between -160 C/A and +54 C/T loci (D'=1) was observed, when compared with -347 G/GA and +54 C/T(D'=0.20) or -347 G/GA and -160 C/A (D'=0.07) loci in cases (Figure 2). In contrast, we observed strong linkage disequilibrium between -347 G/GA and -160 C/A (D'=1) than -347 G/GA and +54 C/T (D'=0.02) or -160 C/A and +54 C/T (D'=0) loci in controls.
Haplotypes
Haplotype frequency
Chi-square
(X2)aP-Valuea
Odd
Ratio95% CI
–347
-160
54
PCOS (%)
Control (%)
G
C
C
76(36.19)
110(50)
Ref.
G
C
T
20(9.52)
8(3.64)
9.193
0.0024
0.2764
0.1157 - 0.6601
G
A
C
24(11.42)
40(18.18)
0.221
0.6360
1.1515
0.6419 - 2.0658
G
A
T
18(8.57)
10(4.55)
5.422
0.0199
0.3838
0.1679 - 0.8771
GA
C
C
25(11.90)
32(14.55)
0.162
0.6873
0.8844
0.4857 - 1.6104
GA
C
T
16(7.61)
14(6.36)
1.644
0.1998
0.6045
0.2786 - 1.3116
GA
A
C
21(10)
5(2.27)
14.638
0.0001
0.1645
0.0594 - 0.4554
GA
A
T
10(4.76)
1(0.45)
10.576
0.0012
0.0691
0.0087 - 0.5511
Ref: Reference; CI: confidence interval;a Fisher’s exact test (2x2 table at 1 df); P<0.05
Table 3: Haplotype frequencies of CDH1polymorphisms in PCOS patients and controls.
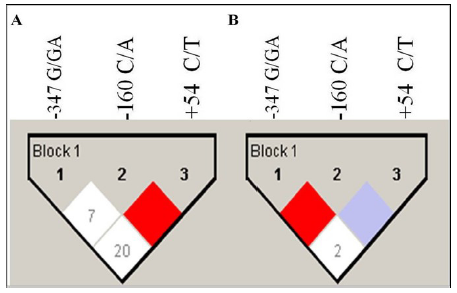
Figure 2: LD analysis of cases and controls are shown separately.
Haploview plots are presented along with the single nucleotide
polymorphisms studied. The pair-wise linkage disequilibrium values
(D’ = 0–100) of all single nucleotide polymorphisms are given
in each diamond. A value of 100 represents maximum possible
linkage disequilibrium. (A) LD analysis of cases. (B) LD analysis of
controls.
Comparison of Minor Allelic Frequency (MAF) of the Studied Polymorphisms from Different Databases
The minor allelic frequency of the studied polymorphisms -347G/GA (rs5030625, GA as minor allele), -160 C/A (rs16260, A as minor allele) and +54 C/T (rs1801026, T as minor allele) were compared with the mutation frequency data from populations of different ethnic origins, obtained from HapMap, 1000Genomes, Genome Aggregation Database (GnomAD) and EXAC database (dbSNP) listed in NCBI (Supplementary Table 2). We observed that the minor allelic frequency found in the cases of the polymorphism rs16260 (MAF=0.3) was close with the values described for Asian population in accordance with the 1000 Genome and GnomAD databases, for rs5030625 (MAF=0.3), the minor allele frequency was found to be close with Africans and Asians whereas the polymorphism rs1801026 (MAF=0.30) showed higher minor allele frequency in the studied Indian population in comparison to all selected populations in the databases.
Polymorphism
Population
Study
HapMap
1000 Genome
GnomAD
EXAC
CDH1
rs5030625Global
-
0.23
0.19
_
European
-
0.11
0.12
_
African
-
0.36
0.33
_
Asian
-
0.23
0.22
_
American
-
0.22
0.19
_
rs16260
Global
0.19
0.23
0.24
-
European
0.27
0.28
0.27
-
African
0.13
0.12
0.16
-
Asian
0.20
0.30
0.29
-
American
0.23
0.24
0.29
-
rs1801026
Global
-
0.15
0.17
_
European
-
0.16
0.15
_
African
-
0.19
0.19
_
Asian
-
0.15
0.16
_
American
-
0.14
0.17
_
Minor allele for rs5030625 - A; rs16260 - A; rs1801026 - T.
Supplementary Table 2: Minor alleles frequencies of the studied polymorphisms from populations of different ethnic origins, obtained from HapMap, 1000 Genomes, GnomA and EXAC database (dbSNP).
Discussion
The underlying mechanism involved in PCOS development is still relatively unknown and SNPs are important tools used for the identification of genetic variations of the disease susceptibility genes [10]. Our study showed an association between all the studied CDH1genepolymorphisms and PCOS.
Epithelial cadherin is a Ca+2-dependent, 120-kDa transmem brane molecule that maintains the integrity of the conjunct cell–cell structure and influences cellular adherence and is expressed in almost all epithelial cells [13]. It is involved in the remodeling of ovarian tissue during the reproductive cycle, which is important for follicle growth, ovulation and luteinization [3,17,22]. The cytoplasmic portion of E-Cadherin binds to a group of connecting proteins called catenins, which mediate the signalling events from the extracellular environment to the interior of the ovarian follicular cells [21]. Genetic variants of the CDH1 gene especially in promoters and 3'-UTR region may regulate gene function and/or transcriptional efficiency [2,23].
The present study showed a significant association between CDH1 -160 C/A, -347 G/GA, +54 C/T polymorphisms and PCOS risk in South Indian women. The -347 G/GA polymorphism is located upstream of the transcriptional start site of CDH1 gene. It has been demonstrated that the -347 GA allele increases the risk of a number of diseases, including endometriosis, bladder cancer, thyroid carcinoma, and breast cancer via down-regulation of CDH1 transcription and reducing E-Cadherin expression when compared to the G-allele [11,12,24,25]. Increased -347 GA allele frequency leads to decreased levels of E-Cadherin that may increase PCOS susceptibility. The -160 C/A polymorphism is located upstream to the transcriptional start site of the CDH1 gene. Previous reports showed that C allele has much higher binding affinity to transcription factor than the A allele [18]. The +54 C/T SNP in 3'-UTR region of a CDH1 gene affect the mRNA stability thereby possibly influence the levels of E-Cadherin [7]. According to the previous studies, C allele decreased the Transcriptional efficiency by 2-fold compared with the T allele [8]. Our study found that individuals harbouring the +54 T allele have a higher risk of developing PCOS compared with those who have the +54 C allele, indicating that the +54 T allele may contribute to the occurrence of PCOS in South Indian women. Interestingly, the present study also revealed that the individuals with CDH1-347/-160/ +54 GCT, GAT, GAAC, and GAAT haplotypes may have a higher risk for development of PCOS, suggesting that the combined effect of the three SNPs on the transcription and expression of CDH1 may be involved in the development and progression of this disease. The MAF of the polymorphisms, rs16260 and rs5030625 found in the cases were close with the Asians represented in 1000 Genome and GnomAD databases, since Indians are a part of Asian ethnic group, whereas, the polymorphism rs1801026 showed higher MAF in comparison to all selected populations in the databases, for further verification, studies with large sample size are needed.
In conclusion this case-control study showed a significant association between the studied polymorphisms and risk of developing PCOS in south Indian women. Further studies with large sample size, expression and functional analysis are required to validate the results presented in this study.
Acknowledgments
We are thankful to all the patients who participated in the present study. Praveen Guruvaiah would like to thank University Grants Commission (UGC), India for awarding Junior Research Fellowship (JRF, NET).
Author’s Contribution
MB conception and design of study, execution of experiments, analysis and interpretation of data, statistical analysis and drafting of manuscript; PG: execution of experiments and data analysis; SS: data analysis; VKV: data analysis; SG: execution of experiments and data analysis; LD: data analysis; MD: acquisition of data; SS: analysis and interpretation of data, drafting of manuscript.
All authors will have seen and agreed to the ‘Author Contribution’ statement.
Funding
This work was supported in part by grants from the SERB (DST), India (Lr. No: SR/FT/LS-188/2009), Department of Science and Technology (DST), India to Prof. Manjula Bhanoori.
Conflict of Interest
The authors declare that there is no conflict of interests.
Ethical Approval
The study was approved by the ethical committee and review board of Centre for Cellular and Molecular biology (CCMB), Hyderabad. In the study all the participants were of South Indian origin (Dravidian linguistic group).
Informed Consent
Informed written consent form was obtained from all subjects prior to participation in this study.
References
- Ajmal N, Khan SZ, Shaikh R. Polycystic Oavary Syndrome (PCOS) and genetic predisposition: A review article. Eur J Obstet Gynecol Reprod Biol. 2019; 3: 100060.
- Al-Moundhri MS, Al-Khanbashi M, Al-Kindi M, Al-Nabhani M, Burney IA, et al. Association of E-cadherin (CDH1) gene polymorphisms and gastric cancer risk. World J Gastroenterol. 2010; 16: 3432-6.
- Baravalle C, Salvetti NR, Mira GA, Pezzone N, Ortega HH. Microscopic characterization of follicular structures in letrozoleinduced polycystic ovarian syndrome in the rat. Arch Med Res. 2006; 37: 830-9.
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21: 263-5.
- Barthelmess EK, Naz RK. Polycystic ovary syndrome: current status and future perspective. Front Biosci (Elite Ed). 2014; 6: 104-119.
- Calcaterra V, Verduci E, Cena H, Magenes VC, Todisco CF, Tenuta E, et al. Polycystic Ovary Syndrome in Insulin-Resistant Adolescents with Obesity: The Role of Nutrition Therapy and Food Supplements as a Strategy to Protect Fertility. Nutrients. 2021; 13: 1848.
- Chen B, Zhou Y, Yang P, Liu L, Qin XP, Wu XT. CDH1 -160C>A gene polymorphism is an ethnicity-dependent risk factor for gastric cancer. Cytokine. 2011; 55: 266-73.
- Deng QW, He BS, Pan YQ, Sun HL, Xu YQ, Gao TY, et al. Roles of E-cadherin (CDH1) genetic variations in cancer risk: a metaanalysis. Asian Pac J Cancer Prev. 2014; 15: 3705-13.
- Elding H, Lau W, Swallow DM, Maniatis N. Dissecting the genetics of complex inheritance: linkage disequilibrium mapping provides insight into Crohn disease. Am J Hum Genet. 2011; 89: 798-805.
- Eric Lai. Application of SNP Technologies in Medicine: Lessons Learned and Future Challenges. 2001; 11: 927–929.
- Govatati S, Tangudu NK, Deenadayal M, Chakravarty B, Shivaji S, Bhanoori M. Association of E-cadherin single nucleotide polymorphisms with the increased risk of endometriosis in Indian women. Mol Hum Reprod. 2012; 18: 280-7.
- Kiemeney LA, van Houwelingen KP, Bogaerts M, Witjes JA, Swinkels DW, den Heijer M, et al. Polymorphisms in the E-cadherin (CDH1) gene promoter and the risk of bladder cancer. Eur J Cancer. 2006; 42: 3219-27.
- Kim HK, Yang Y, Byeon S, Jeong Y, Kwon J, Lee KH, et al. E-Cadherin and Angiopoietin-2 as Potential Biomarkers for Colorectal Cancer With Peritoneal Carcinomatosis. Anticancer Res. 2021; 41: 4497-4504.
- Khan MJ, Ullah A, Basit S. Genetic Basis of Polycystic Ovary Syndrome (PCOS): Current Perspectives. Appl Clin Genet. 2019; 12: 249-260.
- Li Y, Liang J, Kang S, Dong Z, Wang N, Xing H, et al. E-cadherin gene polymorphisms and haplotype associated with the occurrence of epithelial ovarian cancer in Chinese. Gynecol Oncol. 2008; 108: 409-14.
- McAllister JM, Modi B, Miller BA, Biegler J, Bruggeman R, Legro RS, et al. Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci U S A. 2014; 111: E1519-27.
- Machell NH, Farookhi RE. N-cadherin expression and distribution during luteinization in the rat ovary. Reproduction. 2003; 125: 791-800.
- Memni H, Macherki Y, Klayech Z, Ben-Haj-Ayed A, Farhat K, Remadi Y, et al. E-cadherin genetic variants predict survival outcome in breast cancer patients. J Transl Med. 2016; 14: 320.
- Olaniyan OT, Femi A, Iliya G, Ayobami D, Godam E, Olugbenga E, et al. Vitamin C suppresses ovarian pathophysiology in experimental polycystic ovarian syndrome. Pathophysiology. 2019; 26: 331-341.
- Rotterdam ESHRE/ASRM-Sponsored PCOS consensus workshop group. Revised 2003 consensus on diagnostic criteria and longterm health risksrelated to polycystic ovary syndrome (PCOS). Fertil Steril. 2004; 81: 19-25.
- Rowlands TM, Symonds JM, Farookhi R, Blaschuk OW. Cadherins: crucial regulators of structure and function in reproductive tissues. Rev Reprod. 2000; 5: 53-61.
- Ryan PL, Valentine AF, Bagnell CA. Expression of epithelial cadherin in the developing and adult pig ovary. Biol Reprod. 1996; 55: 1091-7.
- Tan M, Xia S, Zhang Q, Zhu J, Bao E. The -160C>a polymorphism in E-cadherin is associated with the risk of nephrolithiasis. PLoS One. 2013; 2: 8.
- Tipirisetti NR, Govatati S, Govatati S, Kandukuri LR, Cingeetham A, Singh L, et al. Association of E-cadherin single-nucleotide polymorphisms with the increased risk of breast cancer: a study in South Indian women. Genet Test Mol Biomarkers. 2015; 17: 494-50.
- Wang YX, Zhao L, Wang XY, Liu CM, Yu SG. Association between E-cadherin (CDH1) polymorphisms and papillary thyroid carcinoma risk in Han Chinese population. Endocrine. 2012; 41: 526- 31.