
Research Article
Austin J Obstet Gynecol. 2021; 8(3): 1174.
Contribution of Pathogenic CNVs and Noonan Syndrome in Fetuses with Increased Nuchal Translucency and Persistently Increased Nuchal Fold
Lin Y1#, Wang H2#, Chau MHK3,4, Lou J1, Zeng X1, Liang Y1, Meng Z2, Zhang R2, Xie R1, Zhong W1, Zhang W5,6, Liu Y1, Yu F1, Choy KW3,4* and Zhu Y2*
1Dongguan Maternal and Child Healthcare Hospital, China
2Department of Obstetrics & Gynaecology, Jinan University, China
3Department of Obstetrics & Gynaecology, The Chinese University of Hong Kong, China
4Shenzhen Research Institute, The Chinese University of Hong Kong, China
5Amcare Genomics Lab, Guangzhou, China
6Department of Molecular and Human Genetics, Baylor College of Medicine, USA
#These authors contributed equally
*Corresponding author: Kwong Wai Choy, Department of Obstetrics & Gynaecology, The Chinese University of Hong Kong, Hong Kong, China; Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, China
Yuanfang Zhu, Maternal-Fetal Medicine Institute, Department of Obstetrics & Gynaecology, Shenzhen Baoan Women’s and Children’s Hospital, Jinan University, Guangdong, China
Received: February 03, 2021; Accepted: March 22, 2021; Published: March 29, 2021
Abstract
Prenatal genetic diagnosis in euploid fetuses with increased Nuchal Translucency (NT) and persistently increased Nuchal Fold (NF) is challenging. The aims of this study is to evaluate the prevalence of pathogenic copy number variants and Noonan Syndrome (NS) in fetuses with increased NT and persistently increased Nuchal Fold (NF) to provide recommendations for pre-natal diagnostic strategies. This is a prospective study from 118 prenatal samples from fetuses with increased NT (≥3.5mm) in first trimester. Multiplex Ligation Dependent Probe Amplification (MLPA), Chromosomal Microarray Analysis (CMA) and karyotyping were conducted. For fetuses with increased NF (≥6mm) in the second trimester with normal karyotype and CMA, targeted Next-Generation Sequencing (NGS) tests for NS were carried out. A total of 118 fetuses had an NT measurement of ≥3.5mm performed MLPA and karyotyping, 89 euploid fetuses were further investigated with CMA, which yielded eight pathogenic CNVs (size ranged from 0.85Mb to 14.5Mb). Twenty fetuses had persistently increased NF at the second trimester. NS testing revealed 3/20 (15%) fetuses had pathogenic variants, and one (5%) with a novel variant of uncertain clinical significance inherited from the father. Our study suggested that NS targeted sequencing facilitates additional genetic diagnosis in fetuses with high NT and persistently increased NF.
Keywords: Nuchal translucency; Nuchal fold; Noonan syndrome; Next generation sequencing; Prenatal diagnosis
Introduction
In prenatal screening it is not uncommon to observe an altered venous-lymphatic differentiation of the endothelial cells in the jugular lymphatic sacs to occur in the first trimester, causing nuchal edema beginning in the second trimester [1,2]. Fetal increased Nuchal Translucency (NT) is recognized as a sensitive marker for chromosomal disorders, and has been applied in routine first trimester Down syndrome screening programs in many countries [3,4], first used to screen for DS by Professor Nicolaides [5]. In the second trimester, about one-third of DS fetuses have a Nuchal Fold (NF) thickness >6mm [6]. NF is the most frequently investigated marker for fetal chromosomal abnormalities [7,8]. Increased NT and increased NF are also associated with many genetic disorders, structural abnormalities and syndromes [9,10].
Chromosomal Microarray Analysis (CMA) has significant advantages over karyotyping in both the prenatal and postnatal genetic diagnosis. CMA is rapidly becoming the preferred diagnostic test for fetuses with structural malformations [11-13]. Pathogenic chromosomal Copy Number Variants (CNVs) can be diagnosed by CMA accurately in fetuses with NT >99th percentile (≥3.5mm) [14,15]. A meta-analysis by M. Grande et al included 17 studies showed the incremental yield of CMA in detecting pathogenic CNVs after a normal karyotype was 5%, although CMA cannot detect balanced rearrangements and single gene mutations [15].
Noonan Syndrome (NS:MIM163950) is regarded as the single gene disorder which is the most frequently associated with increased NT, with an incidence of up to 7% in the first trimester, and 10% in the second trimester pregnancy [16-19]. NS is an autosomal dominant disorder, with a prevalence of 1 in 1000-2500 [20]. This disease is characterized by distinct craniofacial dysmorphisms, postnatal growth retardation, and congenital cardiac defects such as pulmonary valve stenosis, atrial septal defects, and hypertrophic cardiomyopathy. Craniofacial features include broad forehead, hypertelorism, downslanting palpebral fissures, ptosis, and lowset posteriorly rotated ears [21]. NS is a genetically heterogeneous disease, more than 10 genes are associated with NS. Three major genes, including PTPN11 (~50%; MIM 176876) [22], SOS1 (10-15 %; MIM 182530) [23] and RAF1 (5-10%; MIM 164760) [24,25] are known to be causative genes with higher detection rates for NS. The other disease causing genes are KRAS (MIM: 190070), LZTR1 (OMIM 600574), SOS2 (MIM 601247), CBL (OMIM 165360), RIT1 (OMIM 609591), and RRAS (MIM 165090). Molecular diagnostic techniques have developed rapidly, such as targeted Next Generation Sequencing (NGS) tests to enable accurate, faster and cost-effective diagnosis of NS [26]. Bakker M et al. proposed a cost-effective selection of clinical options [27]. They showed that the diagnosis of NS can be suspected prenatally in fetuses with increased nuchal translucency and one or more of the following characteristics: persistently increased nuchal fold or cystic hygroma, hydrops fetalis, pleural effusion, cardiac anomalies, polyhydramnios. At present, managing pregnancy with increased NT involves continuing assessment to establish a proper diagnostic strategy, and to provide parents with realistic information about fetal outcomes. The aims of this study is to evaluate the prevalence of copy number variants and NS in unique cohort of fetuses with increased Nuchal Translucency (NT) and persistently increased Nuchal Fold (NF) to provide data for prenatal diagnostic strategies.
Materials and Methods
Patients and samples
From January 2015 to October 2017, study subjects were referred to two prenatal diagnosis centers for increased NT investigation. The doctors who obtained NT measurement were certified by The Fetal Medicine Foundation, London, UK. NT measurements were performed between gestational ages of 11-13+6 weeks. The NF was measured on a frontooccipital transverse view, including the cavum septum pellucidum, cerebellum and the posterior fossa, as the distance between the median point of the outer curve of the occipital bone and the outer skin edge in 15-20 weeks, the cut-off value is 6mm [28]. 118 pregnancies with NT≥3.5mm were recommended to receive prenatal diagnostic testing in Dongguan Maternal and Child Health Care Hospital and Shenzhen Bao’an Maternal and child health hospital.
Multiplex ligation dependent probe amplification
Multiplex Ligation Dependent Probe Amplification (MLPA) analysis were performed using the SALSA, RMLPA Rprobemix P095 aneuploidy assay (MRC-Holland, Amsterdam, the Netherlands). MLPA assays were used to assess copy numbers of chromosome 13, 18, 21, X and Y. MLPA results were verified by karyotype analysis and CMA test.
Chromosomal Microarray Analysis (CMA)
Genomic Copy-Number Variants (CNVs) were detected using the Fetal DNA Chip (Agilent Technologies, Inc. Santa Clara, CA, USA), a targeted high-resolution 8X60k oligonucleotide array (Fetal DNA Chip), specifically constructed for prenatal diagnosis with the intention of targeting common trisomic aneuploidies and most well-known micro-deletion/microduplication syndromes, or the Affymetrix CytoScan 750k array.
The inclusion criteria for Noonan testing
The inclusion criteria to carry out NGS for NS were: 1. NT measurement ≥3.5mm in the first trimester and NF≥6mm in the second trimester; 2. Normal karyotype and CMA results in the prenatal chorionic villus sampling or amniocentesis.
Noonan testing
DNA extracted from the invasive samples were sent to a diagnostic laboratory to detect NS genes by target capture and sequencing including BRAF, HRAS, KRAS, MAP2K1, MAP2K2, PTPN11, RAF1, SHOC2, SOS1, CBL, NRAS, RIT1. Results with known pathogenic variants in any of the tested genes were considered positive, while those with variants of uncertain clinical significance were considered VUS. No clinically significant variants detected are considered negative. The positive results were confirmed by sanger sequencing.
Target capture and sequencing
Target capture and sequencing: The genomic DNA of the samples was fragmented by a Q800R Sonicator (Qsonica) to generate 300-500bp insert fragments. The paired end libraries were prepared following the Illumina library preparation protocol. Custom designed NimbleGen SeqCap probes (Roche NimbleGen, Madison, Wis) were used for in-solution hybridization to enrich target sequences. Enriched DNA samples were indexed and sequenced on a NextSeq500 sequencer (Illumina, San Diego, Calif) with 100-150 cycles of single end reads, according to the manufacturer’s protocols.
Variant annotation and interpretation: Primary data were received in fastq format after image analysis and base calling conducted using the Illumina Pipeline. The data were filtered to generate ‘clean reads’ by removing adapters and low quality reads (Q20). Sequencing reads were mapped to the reference human genome version hg19 (2009-02 release, http://genome.ucsc.edu/). Nucleotide changes observed in aligned reads were called and reviewed by using NextGENe software (SoftGenetics, State College, Pa). Beside detection of deleterious mutations and novel single nucleotide variants, coverage-based algorithm developed in-house, eCNVscan, was used to detect large exonic deletions and duplications. The normalized coverage depth of each exon of a test sample was compared with the mean coverage of the same exon in the reference file to detect Copy Number Variants (CNVs). Sequence variants were annotated using population and literature databases including the 1000 Genomes Project, dbSNP, GnomAD, Clinvar, HGMD and OMIM. Some online software were used to analyze the structure of the protein, predict the conservation domain, function domain and perform the multiple sequence alignment. Variant interpretation was performed according to the American College of Medical Genetics and Genomics (ACMG) guidelines. The study was approved by the research ethics committee of the institutions.
Results
A total of 118 fetuses had NT measurements of NT ≥3.5mm had MLPA and karyotyping with either CVS or amniocentesis samples detected 29 aneuploidies (Figure 1). The remaining 89 euploid fetuses performed CMA and found eight pathogenic CNVs and five VUS (Table 1 and Table 2). Twenty euploid fetuses had persistently increased NF (≥6mm) in the 15-20 gestational week ultrasound scan with normal CMA results and were offered Noonan Syndrome (NS) gene panel testing. The NS gene panel yielded positive findings in three fetuses (15%, 3/20), a variant of uncertain clinical significance in one (5%, 1/20), and negative results in 16 (80%, 16/20) fetuses (Table 3). The NT/NF thickness measurements of fetuses with NS positive gene panel results were 8mm/11.5mm (case 1), 5mm/10mm (case 2), 5.2mm/8mm (case 3) (Table 3). Fetuses with positive NS variants had a thicker median NT and NF than those with negative results (Table 4). There were no other ultrasonographic structural abnormalities detected in any of the fetuses tested positive for NS. Two fetuses carried known pathogenic mutations in the PTPN11 gene, the other with a known pathogenic mutation in the BRAF gene. Case 1 was found with polyhydramnios. All positive NS results and VUS were confirmed by Sanger sequencing. The fetuses tested positive for NS were terminated, and case 4 with a VUS in BRAF (Table 4) resulted in a live birth. Follow up at the age of ~2 years old revealed mild autistic features. In the remaining 16 fetuses with negative NS results, 3 fetuses had prenatal ultrasound abnormalities. There was one fetus with hydrocephalus, one with hypoplastic left heart, and one with tetralogy of fallot, cerebellar dysplasia and micrognathia. In these three cases, the parents opted for termination of the pregnancy in the second trimester. Follow up of remaining cases reported live birth and healthy babies in all 13 cases (Supplementary Table 1).
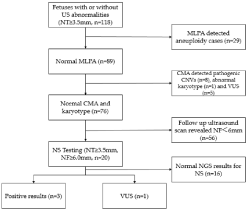
Figure 1: Summary of prenatal testing of fetuses with increased nuchal
translucency.
NS: Noonan Syndrome Testing; CMA: Chromosomal Microarray Analysis;
CNV: Copy-Number Variant; MLPA: Multiplex Ligation-Dependent Probe
Amplification; NT: Nuchal Translucency; NGS: Next-Generation Sequencing;
VUS: Variants of Uncertain Clinical Significance; NF: Nuchal Fold.
Case
NT (mm)
NF (mm)
Findings on US (second trimester)
CNVs resultsa
CNV type
Size (Mb)
Inheritance
Karyotype results
Outcome
1
7
NA
NA
arr 4p16.3 (68345_3449602) x 1
Del
3.3
de novo
Normal
TOP
2
3.5
6.9
Micrognathia, microtia
arr 22q11.21 (18909032_21307800) x 3
Dup
2.4
de novo
Normal
TOP
3
4.1
5.3
No structural abnormality
arr 13q14.2q14.3 (49301719_53625881) x 1
Del
4.3
NA
Normal
TOP
4
3.7
5.6
Tetralogy of fallot
arr 22q11.21 (18909032_21357982) x 1
Del
2.4
de novo
Normal
TOP
5
3.8
5.3
Defect endocardial cushion
arr 16p13.11p12.3 (14,892,975-16,858,332) x 1
Del
1.97
de novo
Normal
TOP
6
3.8
6.5
No structural abnormality
Del
0.85
de novo
Normal
TOP
7
6.3
8.8
Cystic hygroma
arr 5p13.2q11.1 (34985928_49441945) x 3
Dup
14.5
de novo
Abnormal
TOP
8
3.5
7.5
No structural abnormality
arrXp22.31 (6455151_8143509) x 0
Del
1.7
de novo
Normal
TOP
NT: Nuchal Translucency; NF: Nuchal Fold; US: Ultrasound Scan; CNVs: Copy Number Variations; a: UCSC Genome Browser hg19 coordinates; NA: Not Available; TOP: Termination of Pregnancy.
Table 1: Molecular and clinical characterization of 8 fetuses with pathogenic CNVs.
Case
NT (mm)
NF (mm)
Findings on US (second trimester)
CMA resultsa
CNV type
Size (Mb)
Inheritance
Karyotype results
Outcome
1
6.4
NA
Cystic hygroma
arr [GRCh37] 2q14.3 (126,978,133-127,743,822) x1
Del
0.766
Mat
Normal
TOP
arr [GRCh37] 7p22.2 (2,955,638-3,704,743) x 3
Dup
0.749
2
3.6
5.4
No structural abnormality
arr [GRCh37] 22q11.21 (18,970,561-21,800,471) x 3
Dup
2.83
Mat
Normal
LB
3
5.9
7.1
Ventricular septal defect
arr [GRCh37] 7q33 (136,578,081-137,248,769) x 1
Del
0.671
de novo
Normal
TOP
arr [GRCh37] 13q31.1 (80,760,920-82,446,069) x 1
Del
1.68
4
3.7
7
No structural abnormality
arr [GRCh37] 13q12.12 (23,532,645-24,870,530) x 3
Dup
1.3
Pat
Normal
LB
5
3.5
7.5
No structural abnormality
arr [GRCh37] 18p11.32 (179,030-1,900,396) x 1
Del
1.7
Mat
Normal
LB
NT: Nuchal Translucency; NF: Nuchal Fold; US: Ultrasound Scan; Conc: Conclusion; VOUS: Variants of Unknown Significance; CNVs: Copy Number Variations; a: UCSC Genome Browser hg19 coordinates; NA: Not Available; TOP: Termination of Pregnancy; LB: Live Birth.
Table 2: Molecular and clinical characterization of 5 fetuses with VUS CNVs.
No. of fetuses
Maternal age at EDC (Yr)
Median NT thickness (mm)
Median NF thickness (mm)
Positive NS
3
32
6.1
9.8
Negative NS
16
30
4.2
7.1
VOUS
1
33
4.1
6.7
NT: Nuchal Translucency; NF: Nuchal Fold; NS: Noonan Syndrome; VOUS: Variants of Unknown Significance.
Table 3: NT/NF thickness measurements of the fetuses with NS positive gene panel results.
Discussion
In this prospective study, 15% (3/20) of euploid fetuses with increased NT and persistently increased NF and normal CMA were diagnosed genetically with NS using targeted next-generation sequencing. This information will be helpful for clinicians and parents to assess the risks for pregnancy management and future pregnancies. In this study, we describe an increased incidence of pathogenic CNVs (8.98%, 8/89) among the euploid fetuses with increased NT which is similar to the literature [14,29].
We chose the cut-off value of increased NT as 3.5mm according to the 99th percentile of the normal range [9]. Meanwhile, we chose persistently increased NF as a necessary indicator for NS testing based on the literature that this marker is associated with aneuploidies and genetic syndromes [8,10] but it is challenging in prenatal diagnosis. There are no data about the relevance of NS in increased NT and persistently increased NF with normal CMA in literature. In euploid fetuses, increased NT (≥3.0mm) is associated with a high risk 10% (4/39) of NS [30], our study also indicated that NT ≥3.5mm and NF ≥6mm showed a similar risk (15%, 3/20) of NS. Taken together, NS using targeted next-generation sequencing should be recommended for this group of fetus with normal CMA results.
Croonen EA et al. [31] recommend genes including PTPN11, RAF1 and KRAS to be tested when increased NT is combined with one or more of polyhydramnios, hydrops fetal, cystic hygroma, cardiac anomalies, hydrothorax, the detection rate of NS was 17%. Targeted NGS could allow a rapid, less costly and an accurate diagnosis for Noonan syndrome [26]. A targeted NGS for Noonan syndrome could provide further information about this monogenic disease to parents for genetic counseling and decision-making. In our study, 12 targeted genes which were known to be associated with NS were tested, including BRAF, HRAS, KRAS, MAP2K1, MAP2K2, PTPN11, RAF1, SHOC2, SOS1, CBL, NRAS, RIT1 [26,32].
In the NS positive cases, case 1 had an increased NT (8mm) and NF (11mm), but no obviously ultrasound structural abnormalities were found except polyhydramnios until 23 gestation weeks. The positive case 2 (NT: 5.0mm/NF: 10.0mm) and case 3 (NT: 5.2mm/ NF: 8mm) also had no ultrasound structural abnormalities (Table 4). The features of NS include craniofacial features, common heart malformations such as pulmonary valve stenosis, atrial septal defects and hypertrophic cardiomyopathy which are not easily detectable in first trimester or early second trimester ultrasound scans. In our study, all pathogenic NS cases had increased NT and persistently increased NF without prenatal severe structural abnormalities. Consequently, if increased NT is detected by prenatal ultrasound, especially when NF is increased persistently, targeted capture and sequencing for NS or even whole exome sequencing is suggested to rule out NS or other single gene disorders.
Case
NT
NF
Findings on US
NS Results
Conc.
Outcome
1
8
11.5
Polyhydramnios
PTPN11 c.184T>G (p.Y62D)
Likely Path.
TOP
2
5
10
No structural abnormality
PTPN11 c.1528C>G (p.Q510E)
Path.
TOP
3
5.2
8
No structural abnormality
BRAF c.1411G>T (p.V471F)
Path.
TOP
4
4.1
6.7
No structural abnormality
BRAF c.1120A>G (p.I374V)
Vus.
LB
NS: Noonan Syndrome; VUS: Variants of Unknown Significance; TOP: Termination of Pregnancy; LB: Live Birth.
Table 4: Molecular and clinical characterization of 3 fetuses with positive NS gene panel results and 1 VUS.
One fetus carried a VUS in BRAF c.1120A>G (p.I374V), which was inherited from her father. The father showed no abnormalities from detailed physical and ultrasound examination by a geneticist. However, follow up of this child at the age of ~2 years old revealed mild autistic features, including poor verbal skills and attention. Although the penetrance of NS is complete, NS is also known to show variable expressivity. Further follow up assessments of both father and child including cognitive, physical examination and cardiovascular examinations are warranted to investigate whether there is causative association.
In the 16 fetuses with negative NS panel results, all achieved a healthy live birth. Follow up available in 14 cases showed healthy children. The remaining two were lost to follow up. This indicates when NS is excluded, fetuses with increased NT and persistently increased NF have good outcomes. At present, managing pregnancies with increased NT involves continuous assessment to establish a proper diagnostic strategy. Pregnant woman can undergo the CMA as a first-tier test in fetuses with increased NT or, alternative, after common aneuploidies have been ruled out by normal MLPA or QF-PCR results. These rapid molecular diagnostic techniques can be performed immediately after screening in first trimester or in the early second trimester. Fetuses with a normal CMA and karyotype are recommended to perform screening 2-3 weeks later to assess the structural anomalies such as cardiac defects, skeletal deformity and so on, that are known to be associated with increased NT [10], and the measurement of NF is also important for risk assessment. Detailed scan at 20 weeks of gestation is essential to assess the fetal anatomy. When the NF remains increased, the risk of an unidentified anomaly or genetic syndrome remains. In these cases, Noonan testing results could provide parents with realistic information about fetal outcome and help them make informed decisions. With further research and development of whole exome sequencing tests [33], studies with large sample sizes to detect more single gene disease among increased NT fetuses with normal karyotype and chromosomal microarray are needed [34].
The limitation of this study is the small sample size of the increased NT and persistently increased NF, with the reduced cost of WES. A larger sample size study using WES should be conducted in the future.
Conclusion
In summary, we describe an increased rate of pathogenic CNVs (8.98%, 8/89) among euploid fetuses with increased NT. Target capture sequencing for NS yielded an addition-al 15% (3/20) diagnostic rate among fetuses with normal CMA, increased NT and persistently increased NF. Our study suggested that CMA and targeted sequencing of NS genes facilitates genetic diagnosis and enables more accurate prediction of fetal prognosis and genetic risk of recurrence in future pregnancies.
Author Contributions
Yangyang Lin, Huilin Wang and Zhu Yuanfang designed the research, Yangyang Lin, Huilin Wang, Matthew Hoi Kin Chau, Jiwu Lou, Xiumei Zeng, Yuanhao Liang, Zhuo Meng, Rui Zhang, Rungui Xie, Wenbin Zhong, Yanhui Liu, Fubing Yu, Yuanfan Zhu performed research; Huilin Wang, Wei Zhang analyzed data; Yangyang Lin and Huilin Wang, wrote the paper, Yangyang Lin, Huilin Wang, Matthew Hoi Kin Chau and Kwong Wai Choy revised the paper.
Funding
This research was funded by Social Developmental Project of Dongguan City, Guang-dong, China, grant number 2014108101016 and NSFC Emergency Management Project, grant number 81741004, ShenZhen Bao’an Science and Technology Project 2020JD440, Shenzhen Key Medical Discipline Construction Fund (SZXK028).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee) of Dongguan Maternal and child healthcare hospital.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
References
- Bekker MN, van den Akker NM, Bartelings MM, Arkesteijn JB, Fischer SG, Polman JA, et al. Nuchal edema and venous-lymphatic phenotype disturbance in human fetuses and mouse embryos with aneuploidy. J Soc Gynecol Investig. 2006; 13: 209-216.
- Bekker MN, van den Akker NM, de Mooij YM, Bartelings MM, van Vugt JM, Gittenberger-de Groot AC. Jugular lymphatic maldevelopment in Turner syndrome and trisomy 21: different anomalies leading to nuchal edema. Reprod Sci. 2008; 15: 295-304.
- Snijders RJ, Noble P, Sebire N, Souka A, Nicolaides KH. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10-14 weeks of gestation. Fetal Medicine Foundation First Trimester Screening Group. Lancet. 1998; 352: 343-346.
- Leung TY, Chan LW, Law LW, Sahota DS, Fung TY, Leung TN, et al. First trimester combined screening for Trisomy 21 in Hong Kong: outcome of the first 10,000 cases. J Matern Fetal Neonatal Med. 2009; 22: 300-304.
- Nicolaides KH, Azar G, Byrne D, Mansur C, Marks K. Fetal nuchal translucency: ultrasound screening for chromosomal defects in first trimester of pregnancy. BMJ. 1992; 304: 867-869.
- Benacerraf BR, Frigoletto FD, Jr. Soft tissue nuchal fold in the secondtrimester fetus: standards for normal measurements compared with those in Down syndrome. Am J Obstet Gynecol. 1987; 157: 1146-1149.
- Maymon R, Zimerman AL, Weinraub Z, Herman A, Cuckle H. Correlation between nuchal translucency and nuchal skin-fold measurements in Down syndrome and unaffected fetuses. Ultrasound Obstet Gynecol. 2008; 32: 501-505.
- Odibo AO, Sehdev HM, Gerkowicz S, Stamilio DM, Macones GA. Comparison of the efficiency of second-trimester nasal bone hypoplasia and increased nuchal fold in Down syndrome screening. Am J Obstet Gynecol. 2008; 199: 281 e1-5.
- Souka AP, Von Kaisenberg CS, Hyett JA, Sonek JD, Nicolaides KH. Increased nuchal translucency with normal karyotype. Am J Obstet Gynecol. 2005; 192: 1005-1021.
- Ayras O, Tikkanen M, Eronen M, Paavonen J, Stefanovic V. Increased nuchal translucency and pregnancy outcome: a retrospective study of 1063 consecutive singleton pregnancies in a single referral institution. Prenat Diagn. 2013; 33: 856-862.
- Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012; 367: 2175-2184.
- de Wit MC, Srebniak MI, Govaerts LC, Van Opstal D, Galjaard RJ, Go AT. Additional value of prenatal genomic array testing in fetuses with isolated structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature. Ultrasound Obstet Gynecol. 2014; 43: 139-146.
- Chau MHK, Cao Y, Yvonne Kwok KY, Chan S, Chan YM, Wang H, et al. Characteristics and mode of inheritance of pathogenic copy number variants in prenatal diagnosis. Am J Obstet Gynecol. 2019; 221: 493.
- Leung TY, Vogel I, Lau TK, Chong W, Hyett JA, Petersen OB, et al. Identification of submicroscopic chromosomal aberrations in fetuses with increased nuchal translucency and apparently normal karyotype. Ultrasound Obstet Gynecol. 2011; 38: 314-319.
- Grande M, Jansen FA, Blumenfeld YJ, Fisher A, Odibo AO, Haak MC, et al. Genomic microarray in fetuses with increased nuchal translucency and normal karyotype: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2015; 46: 650-658.
- Adekunle O, Gopee A, el-Sayed M, Thilaganathan B. Increased first trimester nuchal translucency: pregnancy and infant outcomes after routine screening for Down’s syndrome in an unselected antenatal population. Br J Radiol. 1999; 72: 457-460.
- Hiippala A, Eronen M, Taipale P, Salonen R, Hiilesmaa V. Fetal nuchal translucency and normal chromosomes: a long-term follow-up study. Ultrasound Obstet Gynecol. 2001; 18: 18-22.
- Lee KA, Williams B, Roza K, Ferguson H, David K, Eddleman K, et al. PTPN11 analysis for the prenatal diagnosis of Noonan syndrome in fetuses with abnormal ultrasound findings. Clin Genet. 2009; 75: 190-194.
- Pergament E, Alamillo C, Sak K, Fiddler M. Genetic assessment following increased nuchal translucency and normal karyotype. Prenat Diagn. 2011; 31: 307-310.
- Mendez HM, Opitz JM. Noonan syndrome: a review. Am J Med Genet. 1985; 21: 493-506.
- Romano AA, Allanson JE, Dahlgren J, Gelb BD, Hall B, Pierpont ME, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2010; 126: 746-759.
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001; 29: 465-468.
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007; 39: 70-74.
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007; 39: 1007-1012.
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007; 39: 1013-1017.
- Lepri FR, Scavelli R, Digilio MC, Gnazzo M, Grotta S, Dentici ML, et al. Diagnosis of Noonan syndrome and related disorders using target next generation sequencing. BMC Med Genet. 2014; 15: 14.
- Bakker M, Pajkrt E, Mathijssen IB, Bilardo CM. Targeted ultrasound examination and DNA testing for Noonan syndrome, in fetuses with increased nuchal translucency and normal karyotype. Prenat Diagn. 2011; 31: 833-840.
- De Jong-Pleij EA, Ribbert LS, Tromp E, Bilardo CM. Three-dimensional multiplanar ultrasound is a valuable tool in the study of the fetal profile in the second trimester of pregnancy. Ultrasound Obstet Gynecol. 2010; 35: 195-200.
- Zhao XR, Gao L, Wu Y, Wang YL. Application of chromosomal microarray in fetuses with increased nuchal translucency. J Matern Fetal Neonatal Med. 2019; 33: 1749-1754.
- Ali MM, Chasen ST, Norton ME. Testing for Noonan syndrome after increased nuchal translucency. Prenat Diagn. 2017; 37: 750-753.
- Croonen EA, Nillesen WM, Stuurman KE, Oudesluijs G, van de Laar IM, Martens L, et al. Prenatal diagnostic testing of the Noonan syndrome genes in fetuses with abnormal ultrasound findings. Eur J Hum Genet. 2013; 21: 936-942.
- Ramond F, Duband S, Croisille P, Cave H, Teyssier G, Adouard V, et al. Expanding the cardiac spectrum of Noonan syndrome with RIT1 variant: Left main coronary artery atresia causing sudden death. Eur J Med Genet. 2017; 60: 299-302.
- Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel AL, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2018; 20: 645-654.
- Qi Q, Jiang Y, Zhou X, Meng H, Hao N, Chang J, et al. Simultaneous Detection of CNVs and SNVs Improves the Diagnostic Yield of Fetuses with Ultrasound Anomalies and Normal Karyotypes. Genes. 2020; 11: 1397.