Abstract
Blast disease caused by Magnaporthe species is major problem faced by rice and finger millet cultivation across the world. Knowing the genetic diversity and population structure of Magnaporthe species is important for designing blast management strategies, to understand the evolution of virulent pathotypes and basis of host shifts. In this study, we used multi-marker system including Simple Sequence Repeats (SSRs), repetitive DNA based markers (Pot2 and Grasshopper), pathogenicity genes and mating locus to study genetic variability of Magnaporthe species in rice and finger millet ecosystems from southern India. Data from multiple markers revealed high genetic diversity and clustering based on geographical location and host species. This study also revealed two rice specific SSR markers (MGM246 and MGM286) and absence of AVR-Co39 and AVR-Pita1 in rice and finger millet, respectively. Interestingly, our study identified multiple copies of grasshopper repeat elements in rice isolates. This element intruded Magnaporthe population infecting finger millet, after evolution of host-specific forms of Magnaporthe. The molecular data obtained using multimarker system, indicated presence of dynamic Magnaporthe population in location of our study. While the clonal nature is known to predominate in field conditions, active gene flow and sexual recombination cannot be denied in cropping zones where different Magnaporthe host crops are co-cultivated. Recurrent characterization of Magnaporthe populations from such locations will help to keep check on emergence of more virulent pathotypes with broad host spectrum.
Keywords: Magnaporthe; Multi-marker; SSR; Pot2; Grh; Pathogenicity genes
Introduction
Magnaporthe oryzae is an Ascomycetes fungal pathogen, which causes blast disease in rice. The genus Magnaporthe consists of several species, which parasitize a wide spectrum of hosts (>50 grass species) including rice, wheat, barley, finger millet and grasses (Panicum italicum; Cenchrus ciliaris; E. indica and E. tristachya) [1-3]. The blast on rice was first described by Cavara as fungal disease caused by Pyricularia oryzae (teleomorph of Magnaporthe oryzae (Hebert)) in 1891 [2]. In 2002, Couch and Kohn designated Digitaria infecting isolates as M. grisea, which is morphologically indistinguishable to M. oryzae. Sexual recombination in Magnaporthe is controlled by a single MAT locus. Fungus of two opposite mating types, MAT1-1 and MAT1-2 are required to produce fertile structure called as perithecia [4,5]. The sexual stage (teleomorph) of M. oryzae has not been found in nature, however sexual reproduction has been reported in vitro conditions between strains of opposite mating types [6,7]. Himalayan foothills are considered to be the center of origin of the M. oryzae [8-11]. Kumar, et al. (1999) have analyzed M. oryzae populations from Indian Himalayas and reported high genetic diversity, presence of both mating types MAT1-1, MAT1-2 and hermaphrodite strains. More recently various genetic events such as deletion, translocation, duplication of Avr gene, or chromosomal rearrangements of field isolates are reported [12]. These genetic variations lead to genome reshuffling which contribute to genome evolution and continuous emergence of virulent strains. High genetic variability coupled with broad host range makes it an ideal model system to study plantpathogen co-evolution [13].
To understand the Magnaporthe population dynamics, many studies have used repetitive DNA based molecular markers such as MGR 586 [14], Pot2 [15] and Grasshopper [16]. Recently, SSR became the most popular molecular marker for genetic mapping [17] and genetic diversity analysis in fungi [18]. However, there are no reports of comprehensive study of Magnaporthe population using combination of marker systems. In this study, we assessed the Magnaporthe species diversity using multiple marker systems including microsatellites, repetitive DNA elements (Pot2 and Grh), and pathogenicity genes and mating locus. In addition, we tested the efficacy of each marker to select best marker system, which can give maximum information about genetic diversity within and between Magnaporthe populations. This is the first study of rice and non-rice Magnaporthe characterization using multiple marker approach.
Materials and Methods
Design of experiment
We selected ten rice (HR12, Co-39, Tadukan, Tetep, MAS26, MAS946-1, Jaya, Intan, KHP9, Rasi, Karizaddu and 27 IRDLs), ten wheat (B. Yellow, DWR-1006, DWR-162, DWR-185, DWR-2006, DWR-39, Kern, UAS-304, UAS-316 and UAS-415) ten finger millet (GPU26, GPU28, GPU45, GPU48, GPU66, GPU67, Indaf5, Indaf9, KMR204, Uduru Mallige and PR202), eleven foxtail millet (Co-7, ISC1162, ISC1209, ISE-995, Narasimharaya, PS-4, RAU-2, RFM- 14, SIA-326, Srilaxmi and TNAU-59) two from each of kodo millet (GPUK-3 and RBK-155), proso millet (TNAU-145 and TNAU-151), little millet (JK-8 and OLM-203) and barnyard millet (VL-172 and VL-207) varieties and planted them in Mandya, Ponnampet and Bengaluru during monsoon season (July–October 2011-2013) for three consecutive years. The seeds of all host varieties were disinfected to avoid seed borne contamination. Disease symptoms were recorded after 21 days of sowing and symptoms were scored (0- 9 scale) as per the Standard Evaluation System (SES), International Rice Research Institute (IRRI), Philippines.
Diseased leaf sampling and single spore isolation
Magnaporthe infected leaves from rice, finger millet, foxtail millet and grasses were collected in 2011, 2012 and 2013 in rainy season (from August to November) from four locations (Bengaluru, Mandya, Ponnampet and Hyderabad). Single spore isolation from infected lesions was performed using method optimized in our laboratory. The infected lesions were surface sterilized by 0.1% Sodium hypochlorite solution followed by two successive washes with sterile water in aseptic laminar hood. A micro humid condition was created by petriplates, pasting sterilized Whatman filter paper on inner surface of upper lid and lower plate filled with sterile water. The surface sterilized leaf samples were pasted on upper lid of these petri plates and incubated under dark at 28oC in an incubator (Innova42, New Brunswick Scientific, USA) for 2-3 days to induce sporulation. Magnaporthe spores were suspended in sterile water and 20 μl of spore suspension was spread evenly using sterile spreader on 2% Agar (Agar-agar, CAS No. 9002-18-0, Fisher Scientific) plate amended with 2 mg of Kanamycin in 100 ml of medium (Kanamycin Sulfate, K1377-5G, Sigma, USA). The single spore was pinpointed under light microscope and scooped using fine tip of sterile Borosilicate glass Pasteur pipets (Cat No. 13-678-20A, Fisher Scientific). Scooped agar with single spore was transferred to oatmeal agar (OMA, cat # M397-500G, Himedia) and allowed to grow for 3-4 days incubated at 28oC under alternate light and dark conditions. The pure cultures of Magnaporthe thus obtained were stored on filter paper discs at -20°C for long term storage.
Genomic DNA extraction
Magnaporthe isolates were grown in a liquid culture (0.2% yeast extract and 1% sucrose) incubated in a shaker incubator at 28oC at 200 RPM for three days. The fungal mycelium was filtered through sterilized miracloth (cat # 475855, Calbiochem, CA, USA) and grounded in liquid nitrogen using Pestle and Mortar. DNA was isolated as per the protocol [19] and DNA quality was checked in Nanodrop ND2000 (Thermo Scientific, DE, USA).
PCR amplification of SSR and MAT locus
We set up 10 μl volume PCR reactions containing 20 ng of genomic DNA, 1 μl of 10 x buffers, 0.4 μl of 20 mM of dNTPs mix, 0.5 μl of 10 mM of each forward and reverse primers, 0.15 μl of Dream Taq (Fermentas, # EP0712, PA, USA). PCR amplification was performed in 2720 Thermal cycle (Applied Biosystems, Foster city, CA, USA) with initial denaturation temperature at 94°C for 5 minutes and followed by 30 cycles with 30 seconds at 94°C, 30 seconds of annealing temperature (variable as per primer provided in the (Table 1)), 1 minute of 72o°C, final extension for 5 minutes at 72°C. PCR products were resolved on 3.5% low Electroendosmosis (EEO) Agarose gel (Himedia, CAS # 9012-36-6, India) stained with GelRed (cat # 41003-1-10 ml, Biotium) and visualized using gel documentation unit (FlourChem, Alpha Innotech, California, USA).
Locus [motif/NCBI ID]
Expected PCR product size (bp)
Forward primer (5'-->3')
Reverse primer (5'-->3')
Tm (°C)
# of cycles
Reference
Simple Sequence Repeats (SSRs)
MGM12 [(tc)17]
202
TGATTGCGTGATCCTTTCAG
CTCTTTTCGCCAAGATCGAC
60
30
Zheng, et al. 2008
MGM51 [(atac)19]
290
GTAACCAGGCCGTTTCAAGA
GGAGGTTGCAGAAGGACAGA
62
30
Zheng, et al. 2008
MGM193 [(at)21]
221
ATTCCCGTGGCTGTGTATTT
GGTCACCCACCCACCTAGTA
58
30
Zheng, et al. 2008
MGM433 [(ta)16]
184
CCTGGCCCAAGCATTATCTA
ACTTGAAGGGCCGAGTTTTC
58
30
Zheng, et al. 2008
MGM437 [(tct)11]
155
GCCCCTCAATAGATCGTCAA
ACTGCGGCATTTTAACCTGT
62
30
Zheng, et al. 2008
MGM240 [(ga)33]
164
CAAGCCCACCGCTAAAATAA
GCCTGCTTCCGTGGTAGTAT
58
30
Zheng, et al. 2008
MGM246 [(gta)14]
178
CCGGATGTCACCTACCAACT
CCTTGTTTTCCCCCTGTGTA
58
30
Zheng, et al. 2008
MGM372 [(at)16]
170
ACGGCTGAACTGCTGTTTCT
GTCACGTCACATTTGCTTGC
62
30
Zheng, et al. 2008
MGM248 [(at)16]
202
CAAGGCTGGTATCCAAGAGG
CTTGAGGAGGTCGTCGATGT
62
30
Zheng, et al. 2008
MGM266 [(tacc)48]
295
TGTGGTGGGTGATCTTGTTG
ATTCCCGGCGAGAGAGATT
62
30
Zheng, et al. 2008
MGM286 [(ttg)37]
184
CGGCTGTGGTTTAACGATTT
CCATCAGGATCCATGAACAC
58
30
Zheng, et al. 2008
MGM288 [(ttg)14]
164
TTGTCGACGAGTGTCCAAAG
CAGTTACCCCGTCGGTATTG
58
30
Zheng, et al. 2008
Host-specificity genes
PWL-1 [U36923]
518
ACGATTAAGTTTGCCGAACA
CGCCAAATAAAACCCTATTCC
55
30
Our study
PWL-2 [U26313]
675
TATGGTCCCGGGTGATAAAA
CCAGGCATACGTTGGAGAAC
60
30
Our study
PWL-3 [U36995]
834
ACCTGCGAGTAAAAGCCTGA
TGTCTGCACCCCTCTCTCTT
58
30
Our study
PWL-4 [U36996]
527
AGCTGCATCCTCTGGAACAT
TGACCATAGTATCCATCCTATTC
55
30
Our study
Avirulent genes
AVR-Pizt [EU837058]
493
TCCCGTCACTTTCATTCTCC
CGAATTCCAGCCGAAGATAC
58
30
Our study
AVR-Pia [AB498873]
450
TGCACACAACAACCTCCATT
GGAATTTTCGGCAGAAATCA
58
30
Our study
AVR-Pii [AB498874]
536
ATTTATGCAGGCCCAAATCC
TGAAATTCCCGCAATAGTCC
62
30
Our study
AVR-Pik [AB498875]
510
ACTGCCACTCTGCACACATC
GTCAAACCTCCCTACGTTGC
62
30
Our study
AVR-Pita1 [AF207841]
675
AGATTCGCAGGCCTCCGAAA
CCTCCATTCCAACACTAACG
60
30
Our study
AVR-CO39 [AF463528]
474
TGCGATATAATGGCCAAACA
GACCGATCTGTCGGGAAGTA
55
30
Couch, et al. 2005
Mating locus
MAT1-1 [NA]
959
TGCGAATGCCTACATCCTGTACCGC
CGCTTCTGAGGAACGCAGACGACC
68
25
Takan, et al. 2012
MAT1-2 [NA]
801
TCTGCTTGAAGCTGCAATACAACGG
CATGCGAGGGTGCCATGATAGGC
68
25
Takan, et al. 2012
Transposon markers
Pot2 [Z33638]
variable
CGGAAGCCCTAAAGCTGTTT
CCCTCATTCGTCACACGTTC
62
30
George, et al. 1998
Grh [M77661]
variable
CTGGGTGGCGAACTCATTAT
AGCGCGAGATTGTTACGC
60
30
Our study
Table 1: Details of primer sequences used for fingerprinting of Magnaporthe isolates.
PCR amplification of Magnaporthe pathogenicity related genes
The cloned pathogenicity gene sequences (CDS) were downloaded from NCBI (Table 1) and primers were designed for 5’ and 3’ of Untranslated Regions (UTR) by considering optimum primer length (19-23 nts), GC content (50-55%) and melting temperature (Tm) using Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/). PCR reaction was set up as described above and PCR program followed with initial denaturation step at 95oC for 5 minutes, 30 cycles of 95°C for 15 seconds, annealing with variable temperature for each primer for 30 seconds, 72°C for 1 minute and final extension at 72°C for 5 minutes. PCR product was separated on 2% low Electroendosmosis (EEO) agarose (Himedia, CAS # 9012-36-6, India). The banding pattern was scored as present (1) and absent (0). A negative control without template DNA was included in all PCR experiments.
Rep-PCR of Pot2 and Grasshopper
The Pot2 repetitive PCR (rep-PCR) [20] was carried out using outwardly directed primers (Table 1). Magnaporthe grisea (Grh) retroelement sequence [16] was downloaded from NCBI database (NCBI, Genbank accession no. M77661). The outwardly directed primers were designed to upstream and downstream regions. The Pot2 and Grh rep-PCR analyses were performed independently in a 25 μl PCR reaction mixture containing 100 ng of genomic DNA, 1x Buffer (600 mM Tris-SO4 (pH 8.9), 180 mM Ammonium Sulfate), 2 mM of dNTPs mix, 1 mM each of outwardly directed primers, 50 mM MgCl2, 2.5 units of hi-fidelity DNA polymerase (Platinum Taq, cat # 11304-011, Invitrogen) and volume made up with nucleasefree water. The PCR program followed was 94°C for 30 seconds as initial denaturation, then 30 cycles of 94°C for 1 min, 62°C (Pot2), 60°C (Grh) for 30 seconds, 68°C for 5 min and 10 min at 68°C as a final extension. We used 1% Megabase agarose (cat # 161-3110, Bio- Rad) along with 0.5% Agarose Clarifier Additive (Synergel, Cat #. SYN-100, Diversified Biotech, USA) to resolve the PCR products in 1x TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA). Gel was stained with gel red and ran for 24 hours for better resolution of bands at 150 V in electrophoresis tank (Sub-cell Model 192, cat # 170-4507, Bio-Rad). The gel image was captured under Flourchem gel documentation unit and bands were scored as present (1) and absent (0). The details of markers used in this study are presented in (Table 1).
Statistical analyses of allelic diversity and Clustering of Magnaporthe isolates
The Polymorphic Information Content (PIC) value was calculated based on allelic frequencies and each SSR was classified based on PIC value as per the criteria (PIC>0.5 highly informative, 0.5>PIC>0.25 reasonably informative and PIC<0.25 slightly informative) proposed by Botstein, et al. (1980). The DNA bands were scored as ‘1’ for present and ‘0’ for absent for a particular allele. The genetic similarity indices were calculated for all the possible combinations (SSR, Pot2, Grh, SSR+Pot2, SSR+Grh, Pot2+Grh and SSR+Pot2+Grh) using Jaccard’s similarity coefficient. All the calculations were performed with the software package NTSYS-pc2.02i [21]. The clustering was done by Unweighted Pair Group Method with Arithmetic Averages (UPGMA) algorithm. The cladogram was constructed with 1000 bootstraps using DARwin [22] and trees were drawn using Dendroscope V3.2.10 [23]. The goodness of fit of similarity matrix indices were tested with Mantel correspondence test [24] in MxCOMP programme in NTSySpc2.02i software package and degree of fit was inferred (very good fit (0.9≤r), good fit (0.8≤r<0.9), poor fit (0.7≤r<0.8) and very poor fit (r<0.7)). The probability of calculated correlation was estimated based on 1000 random permutations.
Mating assay
The isolates of opposite mating types (MAT1-1 and MAT1-2 isolates) were co-cultured on a rice flour medium (14 gm Rice flour, 2.5 gm yeast extract, 15 gm agar and 1 liter distilled water) and allowed to grow at 28°C for three days [9]. Then these plates were transferred to 20°C incubator with continuous fluorescence light for 25-30 days. The plates were observed for formation of fungal fruiting structure perithecium along the line of contact (mating zone).
Results
Survey of Magnaporthe species complex from southern India
To assess the genetic diversity of Magnaporthe species complex, we selected four geographical regions in southern India representing three crop ecosystems; Rice-Rice (Ponnampet and Indian Institute of Rice Research (IIRR), Hyderabad), Rice-Finger millet (V. C. Farm, Mandya) and Finger millet-Finger millet (University of Agricultural Sciences, GKVK, Bengaluru). Ponnampet is rainforest habitat in western ghat region, located in Coorg district of Karnataka and predominantly local rice varieties are grown under high rainfall condition. It is considered to be a global biodiversity hotspot and also as an international hotspot for rice blast disease. This region has been reported to harbor diverse Magnaporthe population [25,26]. The Indian Institute of Rice Research (IIRR), Hyderabad is a Rice- Rice ecosystem where Magnaporthe is maintained in the rice blastscreening nursery. The Gandhi Krishi Vigyana Kendra (GKVK) is located in North Bengaluru, where finger millet is extensively grown under rainfed ecosystem. The fourth location was V. C. Farm, Mandya, where rice and finger millet are co-cultivated. This region was of more interest since Magnaporthe isolates from rice and finger millet might co-exist in this area.
We planted various host plant species and allowed them for natural infection in Bengaluru, Mandya, Ponnampet and Hyderabad (only rice varieties were planted) during south-west monsoon season (July to October) during 2011-2013. Magnaporthe infection was observed in few varieties of rice, finger millet, foxtail millet and Panicum in Mandya (Table S1), see Supporting Information. In Ponnampet, Magnaporthe infection was observed only on rice, finger millet and Digitaria, whereas blast disease symptom found on rice, finger millet, foxtail millet, Cenchrus, E. indica, E. tristachya and Digitaria in Bengaluru (Table S1), see Supporting Information. Diseased leaf samples were collected from infected host varieties. There was no observable infection on wheat, kodo millet, proso millet, little millet and barnyard millet in Mandya, Ponnampet and Bengaluru.
Non-variability of Magnaporthe isolates from single lesion
To check genetic variability of Magnaporthe in single infected lesion, more than one isolate per lesion was sampled. Totally, 258 isolates were fingerprinted using 12 SSRs, Pot2, Grh, 10 pathogenicity genes and 2 alleles of mating locus. However, there was no variability (Figure 1) observed within the isolates derived from same lesion. Unique (one isolate per lesion) set of 146 isolates from 258 was constituted to reduce the redundancy of isolates for the downstream analyses based on the variety and lesion types. These isolates comprised of 81 Magnaporthe from rice, 39 from finger millet and 26 from grasses (Table S1), see Supporting Information.
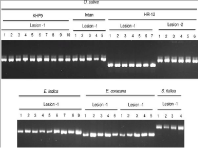
Figure 1: SSR (MGM12) fingerprint profile of Magnaporthe isolates from
single lesion. The host species are represented by their botanical names
(Rice: O. Sativa; Goosegrass: E. Indica; Finger millet: E. Coracana; and
Foxtail millet: S. Italica) and number of monoconidium isolated per lesion are
represented by numbers.
Polymorphic SSRs, allelic richness and diversity in Magnaporthe population
Twelve simple sequence repeats markers (SSRs: 6 di-nucleotide, 4 tri-nucleotide and 2 tetra nucleotide repeats) were chosen based on number of alleles sampled from the previous study [18]. We detected 3 to 8 alleles per SSR locus in Magnaporthe isolates in four locations (Table 2). Eight SSR markers (MGM12, MGM51, MGM193, MGM437, MGM246, MGM240, MGM266 and MGM288) were found to be highly polymorphic (5 alleles) (Table 2). The remaining four SSR markers (MGM433, MGM372, MGM248 and MGM286) were moderately polymorphic (3 to 4 alleles). Interestingly, MGM246 (except two finger millet isolates Ec-M-GPU48-L1-1 and Ec-PGPU26- L1-1) and MGM286 (except four finger millet isolates; Ec- M-GPU48-L1-1, Ec-P-GPU26-L1-1, Ec-P-GPU26-L2-1 and Ec-PGPU67- L1-1) did not amplify in all finger millet and grass isolates (Figure 2) indicating the absence of these locus. But these markers were predominantly present in rice isolates.
Marker
Rice Isolates
Finger millet Isolates
Grass Isolates
All Isolates
No. of alleles
PIC
No. of alleles
PIC
No. of alleles
PIC
No. of alleles
PIC
MGM12
4
0.6
4
0.68
3
0.77
5
0.68
MGM51
6
0.94
3
0.63
7
0.89
8
0.82
MGM193
6
0.67
3
0.15
4
0.75
7
0.52
MGM433
3
0.24
4
0.68
4
0.88
4
0.6
MGM437
7
0.54
3
0.62
6
0.93
7
0.7
MGM246
5
0.64
2
-
3
-
5
0.64
MGM240
3
0.46
4
0.65
3
0.9
5
0.67
MGM372
3
0.35
2
0.35
3
0.88
3
0.53
MGM248
3
0.7
4
0.67
2
0.64
4
0.67
MGM266
7
0.87
1
0.15
3
0.78
8
0.6
MGM286
4
0.56
2
-
0
-
4
0.56
MGM288
7
0.75
2
0.15
2
0.85
8
0.58
Note: No PCR amplification is shown as negative (-).
Table 2: Polymorphic Information Content (PIC) of SSR markers.

Figure 2: SSR fingerprint profile of rice and non-rice isolates for MGM246
and MGM286. Magnaporthe isolates from rice, finger millet, grass are
represented in lane 1-16, 17-21 and 22-27 respectively. The NTC represents
the No Template Control.
The allelic frequency was calculated for all SSR bands and Polymorphic Information Content (PIC) was estimated based on individual host species (rice, finger millet and grasses) or combined populations (rice+finger millet+grasses) across four locations. The combined PIC value for SSR markers was more than 0.5 in our study, which is reported to be more informative [27]. The PIC for individual population was more informative for 9 out of 12 markers in rice, 6 in finger millet and 10 in grasses (Table 2).
In total, 68 alleles were identified in rice, finger millet and grass isolates from 12 SSR markers survey from four locations. Of which, 3 were rare (alleles present in <1% of population), 49 common (alleles present in >1-20% of population), 13 frequent (alleles present in >20- 50% of population) and 3 most frequent (alleles present in >50% of population) alleles. Further comparison of 68 alleles among isolates from different hosts showed those 19 common alleles between rice, finger millet and grasses, 22 rice-specific, 2 finger millet-specific and 1 grass-specific allele(s). Two-way comparisons of alleles showed that 6 alleles were common between rice and finger millet, 7 between finger millet and grass and 11 between rice and grasses (Figure S1, see Supporting Information).
Transposon (Pot2 and Grh) based polymorphism in Magnaporthe species
We used rep-PCR strategy [20] for Pot2 and Grh elements to fingerprint Magnaporthe isolates. We obtained Pot2 rep-PCR amplification for 97.53% of rice, 84.62% of finger millet and 58% grass isolates (Figure 3). Based on rep-PCR amplification pattern, Pot2 elements are closely distributed (distance between Pot2 insertions ranged between 0.6 to 4.5 Kb) on the genomes of rice isolates as compared to finger millet isolates (distance between Pot2 insertions ranged between 0.6 to 7 Kb). The insertion frequency was found to be higher in grass isolates (2-18) as compared to finger millet isolates (2-12).
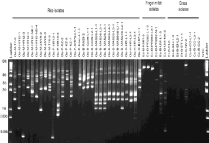
Figure 3: Pot2 fingerprint profile of Magnaporthe isolates collected from
rice, finger millet and grasses. The origin of Magnaporthe isolates are
represented as OS: O. Sativa; Ec: E. Coracana; SI: S. Italic; EI: E. Indica;
ET: E. Tristachya; and CC: Cenchrus Spp. Locations are represented as M:
Mandya; B: Bengaluru; P: Ponnampet; H: Hyderabad; NTC: No Template
Control.
We designed Grasshopper (Grh) rep-PCR strategy to amplify intervening sequences using Grh insertions sites across the Magnaporthe genome. Grasshopper (Grh) is a gypsy like retrotransposon identified in Magnaporthe that infects finger millet and goosegrass [16]. In this study, we identified Grh rep-PCR amplicons for 60% of rice isolates with varied copy numbers ranging from 2 to 14 (Figure 4). Grh rep-PCR amplification was observed for 34% of finger millet isolates and 23% of grass isolates with 2-10 copies. The frequency of insertions varied from 0.35-4 Kb in rice isolates as compared to 0.5-4 Kb in finger millet and grass isolates.
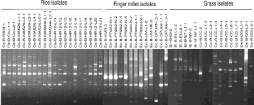
Figure 4: Grasshopper fingerprint profile of Magnaporthe isolates collected
from rice, finger millet and grasses. The origin of Magnaporthe isolates are
represented as OS: O. Sativa; EC: E. Coracana; SI: S. Italic; EI; E. Indica; ET:
E. Tristachya; C: Cenchrus Spp; DS: Digitaria Spp. Locations are represented
as M: Mandya; B: Bengaluru; NTC: No Template Control.
Genetic relatedness of Magnaporthe population
Based on geographical regions: We analyzed the genetic relatedness of Magnaporthe populations from three locations (Mandya, Ponnampet and Bengaluru) by combining data from SSR, Pot2 and Grh. Although, we collected isolates from four locations, Hyderabad was not included in location specific analysis, since we sampled very few rice isolates. The combined analyses from all markers showed two major clades (rice and non-rice) formed at 92% genetic dissimilarity with 24 Magnaporthe lineages in Mandya, out of which, 16 were rice, 6 finger millet, 1 each from foxtail millet and Panicum (Figure 5A). In Ponnampet, overall topology of dendrogram indicated the presence two major clades (rice and non-rice) formed at 91% of genetic dissimilarity with 33 Magnaporthe lineages comprising of 17 rice, 9 finger millet and 7 Digitaria (Figure 5B). Isolates from rice, finger millet and Digitaria formed independent clusters. One of the finger millet isolates (Ec-P-GPU67-L1-1) grouped into rice clade, which formed a separate lineage. There were two major (rice and nonrice isolates) and three minor (finger millet, Digitaria and Cenchrus isolates) clades in GKVK, Bengaluru (Figure 5C). In this location, 30 Magnaporthe lineages were observed, of which 6 from rice, 14 from finger millet, 2 from goose grass (E. indica and E. tristachya), 1 from foxtail millet, 5 from Cenchrus and 2 from Digitaria. The host and cultivar specific clustering was noticed in Bengaluru except for one Cenchrus (Cc-B-CC-2) and one Digitaria (Ds-B-DG-L2-1) isolate, which clustered in finger millet clade. In general we observed that isolates were clustered based on host and cultivar.
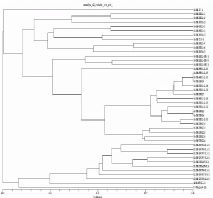
Figure 5a: Dendrogram illustrating the relationships among clonal lineages
of rice and non-rice isolates collected from Mandya revealed by SSR, Pot2
and Grh data. The origin of Magnaporthe isolates are represented as OS:
O. Sativa; EC: E. Coracana; SI: S. Iitalic; PS: Panicum Spp. Location is
represented as M: Mandya; Dotted line indicates 70% similarity cutoff applied
to classify the Magnaporthe isolates into lineages: rice, finger millet and grass
lineages are depicted by dark circle, triangle and square respectively.
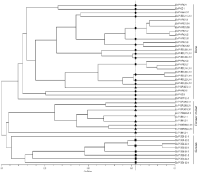
Figure 5b: Dendrogram illustrating the relationships among clonal lineages
of rice and non-rice isolates collected from Ponnampet location revealed by
SSR, Pot2 and Grh data. The origin of Magnaporthe isolates are represented
as OS: O. Sativa; EC: E. Coracana; SI: S. Italic; PS: Panicum Spp. Location
is represented as P: Ponnampet; Dotted line indicates 70% similarity cutoff
applied to classify the Magnaporthe isolates into lineages: rice, finger
millet and grass lineages are depicted by dark circle, triangle and square
respectively.
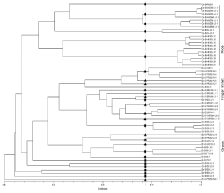
Figure 5c: Dendrogram illustrating the relationships among clonal lineages
of rice and non-rice isolates collected from Bengaluru revealed by SSR,
Pot2 and Grh data. The origin of Magnaporthe isolates are represented as
OS: O. Sativa; EC: E. Coracana; SI: S. Italic; PS: Panicum Spp. Location
is represented as B: Bengaluru; Dotted line indicates 70% similarity cutoff
applied to classify the Magnaporthe isolates into lineages: rice, finger
millet and grass lineages are depicted by dark circle, triangle and square
respectively.
Based on different marker system: The cladogram based on SSR data from four locations grouped rice and finger millet isolates into two distinct major clades (Figure S2, see Supporting Information). Interestingly independent minor clades were formed by 1 finger millet (Ec-B-PR202-N2-1), two Digitaria (Ds-P-DG1-L1-1, Ds-PDG1- L2-2) and 1 Cenchrus (Cc-B-CC-L1-3) isolates (Figure S2, see Supporting Information). Two finger millet isolates (Ec-P-UM-L2-1, Ec-P-GPU67-L1-1) and four Digitaria isolates (Ds-P-DG6-L3-1, Ds-P-DG1-L3-1, Ds-B-DG-L1-1 and Ds-B-DG-L3-1) admixed in the rice clade. Isolates from Cenchrus, Setaria, Digitaria, E. indica and E. tristachya were placed in finger millet clade (Figure S2, see Supporting Information).
The Pot2 based clustering formed a major clade consisting of mixture of isolates from rice, finger millet, Digitaria, Setaria, E. indica, E. tristachya and Cenchrus (Figure S3, see Supporting Information). Few rice (21) and Digitaria (5) isolates clustered together in separate minor clade. The pattern of clustering was random and host-specific or region specific clustering was not observed unlike SSRs. Grh based cladogram also showed random pattern and clustering of isolates (Figure S4, see Supporting Information).
The combined analyses from three marker systems (SSR, Pot2 and Grh) in Magnaporthe revealed two major clades of rice and finger millet that correspond with host-specific nature of the isolates (Figure 6). All Digitaria isolates were grouped in separate cluster within rice clade. One of the finger millet isolates (Ec-P-GPU67-L1-1) was grouped with rice isolate (Os-M-HR12-L25) within rice clade (Figure 6). This indicates that it might be a possible migrant from rice to finger millet host. Location and marker wise clustering revealed host specific and location specific grouping of isolates. Several sub-groups were formed within main clusters indicating the richness of genetic variability within isolates of same host species.
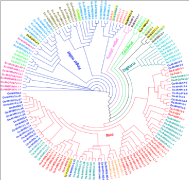
Figure 6: Cladogram (UPGMA) of Magnaporthe strains isolated from Rice,
Finger millet, Foxtail millet, Cenchrus, Digitaria, Goose grass and Panicum
using SSR, Pot2 and Grh data. Edges are represented in red: rice clade;
blue: finger millet; green: Cenchrus; pink: Setaria; dark green: Digitaria;
black: mixed clade. The origin of Magnaporthe isolates are represented as
OS: O. Sativa; EC: E. Coracana; SI: S. Italic; EI: E. Indica; ET: E. Tristachya;
DS: Digitaria Spp; PS: Panicum Species. Locations are represented in M:
Mandya; B: Bengaluru; P: Ponnampet; H: Hyderabad. The isolates shaded in
yellow color are admixtures.
Correlation analysis of genetic similarity indices between markers using Mantel test
We generated genetic similarity matrices for seven possible combinations (SSR, Pot2, Grh, SSR+Pot2, SSR+Grh, Pot2+Grh and SSR+Pot2+Grh) of three marker systems. Then Mantel test [24] was applied to check the goodness of fit of similarity matrices [28,29]. Correlation analysis was performed based on location (Mandya, Ponnampet and Bengaluru) and host (rice and non-rice species), the correlations coefficients are summarized in (Table 3).
Marker
SSR
Pot2
Grh
SSR+Pot2
SSR+Grh
Pot2+Grh
SSR+Pot2+Grh
Location wise correlation
Bengaluru
SSR
-
0.03
-0.02
0.98
0.97
-0.01
0.95
Pot2
-
-
0.02
-0.02
-0.03
0.32
-0.02
Grh
-
-
-
-0.02
-0.03
0.32
-0.02
Mandya
SSR
-
-0.06
-0.04
0.97
0.98
-0.04
0.96
Pot2
-
-
0.22
-0.05
-0.06
0.71
-0.05
Grh
-
-
-
-0.03
-0.03
0.32
-0.03
Ponnampet
SSR
-
0.03
0.26
0.94
0.96
0.29
0.91
Pot2
-
-
-0.03
0.04
0.02
0.04
0.03
Grh
-
-
-
0.24
0.32
0.07
0.29
Host wise correlation
Rice
SSR
-
-0.03
0.06
0.96
0.96
-0.03
0.94
Pot2
-
-
0.12
-0.02
-0.03
1
-0.02
Grh
-
-
-
0.05
0.08
0.12
0.06
Finger millet
SSR
-
0.03
-0.1
0.97
0.97
0.32
0.95
Pot2
-
-
-0.02
0.09
0
-0.04
0.04
Grh
-
-
-
-0.1
-0.09
-0.1
-0.09
Grass
SSR
-
0.11
-0.03
0.85
0.9
0.01
0.81
Pot2
-
-
0.04
0.12
0.02
0.12
0.05
Grh
-
-
-
-0.03
-0.05
0.58
-0.04
All isolates
SSR
-
0.01
0.05
0.96
0.96
-0.02
0.94
Pot2
-
-
0.01
0.03
-0.01
0.23
0.01
Grh
-
-
-
0.05
0.05
0.15
0.05
Table 3: Mantel correlations (r) obtained between the genetic similarities calculated using SSR, Pot2 and Grh. The goodness of fit based on correlations values (r) are interpreted as very good fit (0.9≤r), good fit (0.8≤r<0.9), poor fit (0.7≤r<0.8) and very poor fit (r<0.7).
Location wise analysis revealed that SSR based clustering highly correlated with SSR+Pot2 (<0.90), SSR+Grh (<0.90) and SSR+Pot2+Grh (<0.90). Pot2 or Grh showed very poor fit as per Mantel test with any of the above-mentioned combinations. Hostwise mantel test showed SSR combined with Pot2 or Grh or all three markers together (SSR+Pot2+Grh) was found to be good. Correlation values for all markers of each location and hosts are provided in (Table 3). The trend of correlation between markers remained same across all three locations and also on different hosts’ species.
In summary, we observed better correlation of SSR+Pot2 for classifying rice isolates and SSR+Grh to classify finger millet and grass isolates. Thus SSR alone or combined with either of Pot2 or Grh or both is the best marker to assess the genetic diversity of Magnaporthe population, whereas Pot2 or Grh markers alone are inefficient.
Pathogenicity genes in Magnaporthe populations adapted to different hosts
Pathogenicity genes are the key loci in the Magnaporthe genome, which dictates host specificity and infectivity. In this study, the distribution of host specificity and Avr genes was evaluated using gene-specific PCR amplification in Magnaporthe isolates of rice, finger millet and grass (Figure 7). Interestingly, PWL2 (96.84%), AVR-Pizt (84.81%) and AVR-Pik (68.35%) genes were more frequently present in rice isolates. PWL1 (90%), PWL3 (76%), PWL4 (94%), AVR-Pii (62%) and AVR-Co39 (94%) were predominantly found in finger millet isolates. In case of grass, more than 50% of isolates showed presence of AVR-Pizt, PWL2, PWL3, PWL4, AVR-Co39 and AVRPii. AVR-Co39 was absent in rice isolates and AVR-Pita1 was absent in finger millet isolates.
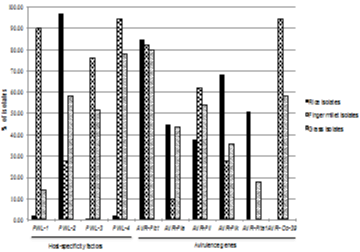
Figure 7: Distribution of pathogenicity genes in Magnaporthe strains isolated
from four geographical regions of India.
Distribution of mating types in Magnaporthe species
Compatibility of sexual recombination between two Magnaporthe isolates is mainly governed by two genes (MAT1-1 and MAT1- 2) of mating locus [4]. Distribution of these alleles in particular Magnaporthe population gives an idea about possibility of mating. In this context, we have analysed distribution of mating types in Magnaporthe population across four locations (Table 4). Over all MAT1-1 was the predominant allele (83.95%) among rice isolates in all four locations. Rice isolates in Mandya (87.50%), Ponnampet (96%) and Hyderabad (100%) were highly skewed towards MAT1- 1 and moderately skewed in Bengaluru (60 %). Rice isolates in Bengaluru, showed higher level of MAT1-2 (40%) as compared to other three locations. This could be due to repeated cultivation of finger millet crop over years in Bengaluru. MAT1-1 (55.56%) and MAT1-2 (44.44%) were equally distributed in finger millet isolates in Mandya, (Table 4) where finger millet and rice crops are sympatrically grown. Finger millet isolates in Ponnampet were skewed towards MAT1-1 (80%) than MAT1-2 (20%), this could be due to extensive cultivation of rice in Ponnampet. In Magnaporthe isolates from grasses (Cenchrus, Digitaria, goose grass, Setaria and Panicum), MAT1-1 (65.38%) is most predominant allele as compared to MAT1-2 (15.38%). Interestingly, MAT locus was not amplified for 3 isolates from Cenchrus and 2 isolates from Digitaria. Study on MAT locus in Magnaporthe isolates from Argentina [30] and Korea [31] have shown that all isolates belong to mating type MAT1-1. The African study revealed that 29% of Magnaporthe isolates belongs to MAT1-1 and 71% to MAT1-2 [32].
Mating locus
Rice Isolates
Finger millet Isolates
All grass Isolates
Ma
Pa
Ba
Ha
Total
Ma
Pa
Ba
Total
MAT1-1 (%)
87.5
96
60
100
83.95
55.56
80
30
48.72
65.38
MAT1-2 (%)
12.5
4
40
0
16.05
44.44
20
70
51.28
15.38
Undeterminedb (%)
0
0
0
0
0
0
0
0
0
19.23
aLocations are represented as M=Mandya, P=Ponnampet, B=Bengaluru and H=Hyderabad.
bIsolates did not amplify either of mating locus.
Table 4: Distribution of mating types in Magnaporthe isolates.
Magnaporthe Teleomorph (sexual stage) stage was induced in laboratory mating experiments by crossing isolates of opposite mating types [33,34]. To know the crossability of Magnaporthe isolates of rice and non-rice hosts, we performed an in vitro mating experiment by co-culturing strains of opposite mating types (Figure 8). We performed 177 combinations (within and between rice and non-rice isolates) using 21 MAT1-1 isolates (5 rice, 8 finger millet and grass each) and 13 MAT1-2 isolates (1 rice, 10 finger millet and 2 grass) (Table S2), see Supporting Information). We observed visible perithecia structure from six isolates combination of E. coracana, E. tristachya, Setaria and Cenchrus [Ec-B-PR202-L2-1 (MAT1-2) x Ec-B-PR202-L1-2 (MAT1-1); Ec-B-Indaf9-L1-1 (MAT1-2) X Ec-BGPU66- L1-1 (MAT1-1); Ec-B-UM-NK-2 (MAT1-2) X Et-B-ET-L1-1 (MAT1-1); Ec-M-Indaf9-L1-1 (MAT1-2) x Ec-B-GPU66-L1-1 (MAT1-1); Ec-M-Indaf9-L1-1 (MAT1-2) X Ec-M-GPU67-N1-1 (MAT1-1); Si-M-NV-L1-1 (MAT1-2) x Ec-B-GPU66-L1-1 (MAT1- 1); Si-M-NV-L1-1 (MAT1-2) x Cc-B-CC-L4-1 (MAT1-1)]. We did not observe mating in any of the rice isolates but fertile strains were found in some of the isolates of finger millet, Setaria, E. tristachya and Cenchrus (Table S2), see Supporting Information.

Figure 8: Mating assay of finger millet isolates. The zone of mating is shown
in white arrow.
Discussion
The blast disease caused by Magnaporthe oryzae is the disastrous biotic stress affecting cultivation of rice, the most important staple food crop of the world. In addition to rice blast, wheat blast and finger millet blast are major concerns in certain regions of Asia and Africa. The use of resistant cultivars and fungicides are commonly deployed measures for control of the blast disease. The use of resistant cultivars is supposed to be environment friendly and sustainable approach; however most of the resistant cultivars become susceptible to blast disease within 2-3 years of release. The main reason behind breakdown of resistance is highly variable genetic nature of Magnaporthe field isolates. Understanding the population dynamics is one of the preliminary steps towards developing region specific disease control strategies. Considering the genome dynamics and continuously evolving nature of Magnaporthe population, we analyzed rice and non-rice Magnaporthe isolates from south India. Our study indicated that, use of combination of multiple markers is an ideal approach to understand population dynamics of Magnaporthe in a given geographical region. The phylogenetic analysis revealed high genetic diversity and independent clustering of Magnaporthe isolates based on host and location. The Simple Sequence Repeats (SSRs) were found to be most informative markers as compared to repetitive DNA based markers Pot2 and Grh. In addition to being more informative, two of the SSR markers MGM246 and MGM286 were specific to rice subpopulation. These markers might be linked to host specificity determinants in rice Magnaporthe population. These markers and the genomic functions of loci adjacent to them should be looked upon to find probable host specificity determinants in rice.
Repetitive DNA sequences have been used as molecular markers to distinguish isolates of Magnaporthe from different host species. The frequently used repeat elements in Magnaporthe are Magnaporthe grisea repeat 586 (MGR 586) [14] and Pyricularia oryzae DNA transposon 2 (Pot2) [15]. MGR586 and Pot2 elements are reported to be multi-locus (~100 copies) with high copy number in rice isolates and low copy numbers or completely absent in non-rice isolates [15,35-37]. We used Grh as another transposable element marker in rep-PCR. Grh (5233bp) is present in multiple copies (>25) in genomes of Magnaporthe isolates from Eleusine spp. This element was reported to be exclusively present in Eleusine isolates of Magnaporthe. However, we found this element in significantly large number of rice Magnaporthe isolates. The presence of these elements in rice isolates can be explained by multiple possibilities, a recent acquisition in rice subpopulation of Magnaporthe or rice Magnaporthe host shift to finger millet. A genetic recombination between rice and finger millet subpopulation in cropping zones could also be the probable reason of wide spread presence of Grh element. Based on efficacy test of markers, we conclude that transposable element based markers alone are not ideal for population studies of Magnaporthe. Combination of transposable element based markers and SSRs or SSRs alone can be used for Magnaporthe population analysis.
We also analyzed the distribution of well-characterized Avr genes in Magnaporthe population. Loss of Avr-Co39 in rice isolates and AVR-Pita1 in finger millet isolates was previously reported [12,38]. We could also observe the similar trend among rice and finger millet isolates. Frequent association of transposons with avirulent genes has been reported in Magnaporthe [39,40]. Failure to amplify AVR-Co39 gene in rice isolates and AVR-Pita1 in finger millet isolates from our study indicates that these genes could have undergone structural rearrangement, complete or partial loss of gene fragment(s), during course of time. The spectrum of avirulence genes in pathogen and resistant genes in host should be analyzed simultaneously and the information gained should be used to screen resistant gene analogues in rice and finger millet. Cognate resistant (R) genes for most prevalent Avr genes in Magnaporthe population can be used to design location specific resistance breeding strategies in rice and finger millet. Breeders can focus on allele mining for these ‘R’ genes and pyramiding of these genes in commonly grown cultivars in South India. Resistance breeding is highly neglected in case of finger millet improvement programme. These necessitates the research on mapping of inherent ‘R’ genes and Resistant Gene Analogues (RGA) of well-characterized ‘R’ genes of rice and deploy them in finger millet resistance breeding. Information from grass-specific Avr genes can be used to map novel ‘R’ genes and look for corresponding RGAs in rice and finger millet. Well-characterized but ineffective ‘R’ genes in rice can be replaced by their respective RGAs from grass families.
The sexual reproduction and mating is one of the proposed mechanism used to generate genetic variability in Magnaporthe population. The role of sexual recombination in generating pathotype diversity in Magnaporthe pathosystem is always in the realm of speculation. So far, the sexual stage of Magnaporthe has not been observed in nature and the field isolates are claimed to follow the asexual life cycle for their propagation. The sexual recombination offers a means of gene flow between different host-specific forms of Magnaporthe resulting in evolution of virulent pathotypes with wider host range. Thus, the sexual recombination has been of wide scientific interest in view of observed pathotype diversity of Magnaporthe isolates. Screening of mating locus alleles, is one of the means of speculating the occurrence of sexual recombination in particular location. We observed the presence of both mating alleles, with MAT1- 1 allele as the more predominant allele in the overall population. In our study, rice isolates showed no crossability, whereas finger millet isolates mated with grass isolates suggesting possible exchange of genetic material among non-rice isolates (E. tristachya, Setaria and Cenchrus). Absence of mating in rice isolates recapitulated the clonal nature of rice isolates. Similar findings leading to evolution of hostspecific forms and emergence of new pathotypes were reported in past [35,36,41-43]. Crossing of Magnaporthe isolates from different hosts may provide clue of evolutionary relationship and mode of reproduction in given location. Evolution of new pathogenic strains and host shifts emphasizes the significance of mating and genetic recombination studies in Magnaporthe disease biology.
Our results suggest that, the population structure can vary drastically and immense genetic variability can be observed among pathogen population. Novel pathotypes can be rendered due to seed exchange and trade. Considering the dynamics of genetic content of a pathogen, recurrent characterization of Magnaporthe population is necessary to design anticipatory resistance breeding in rice and finger millet. Some markers used in this study could clearly distinguish between rice and non-rice isolates, however wide spread presence of the repetitive element grasshopper indicates that there is no clear distinction between rice, finger millet and grass isolates at genetic level. The dissimilarity at genetic level can be attributed to events of sexual recombination or horizontal gene transfer in rice and non-rice Magnaporthe population; however further studies are required in this area. Our recent report on whole genome sequencing and comparative genomics of rice and non-rice Magnaporthe isolates revealed hostspecific secretory proteins and host specific as well as random distribution of Transposable Elements (TEs) and their insertion sites at genome scale[44,45]. Multiple environmental stress factors might be directing the evolution of highly variable, virulent pathogen population with expanded host boundaries. The genetic structure and dynamics of pathogen population, is subjective to multiple factors like mutations, climate variation, selection forces, presence of genetic recombination, gene flow and genetic drift due to introduction of new gene pool. While the clonal nature is known to predominate in field conditions, active gene flow and sexual recombination cannot be denied in cropping zones where different Magnaporthe host crops are co-cultivated. Thus, it is necessary to extrapolate blast pathogen population surveys to major rice and finger millet growing areas. Collectively, our study has generated valuable resources to accelerate blast research in many aspects. This information can be used for generating sustainable disease control, understanding of genomic factors involved in pathogenesis, evolutionary mechanisms and host specificity.
References
- Maciel JLN, Ceresini PC, Castroagudin VL, Zala M, Kema GHJ, McDonald BA. Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology. 2013; 104: 95-107.
- Ou SH. Rice diseases. Philippines: IRRI. 1985.
- Sweigard JA, Carroll AM, Kang S, Farrall L, Chumley FG, Valent B. Identification, cloning and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 1995; 7: 1221-1233.
- Kang S, Chumley FG, Valent B. Isolation of the mating-type genes of the phytopathogenic fungus Magnaporthe grisea using genomic subtraction. Genetics. 1994; 1382: 289-296.
- Valent B, Chumley FG. Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu Rev Phytopatho. l 991; 29: 443-467.
- Hayashi N, Li CY, Li JL, Naito H. In vitro production on rice plants of perithecia of Magnaporthe grisea from Yunnan, China. Mycol Res. 1997; 101: 1308-1310.
- Silue D, Notteghem J. Production of perithecia of Magnaporthe grisea on rice plants. Mycol Res. 1990; 94: 1151-1152.
- Kumar J, Nelson RJ, Zeigler RS. Population structure and dynamics of Magnaporthe grisea in the Indian Himalayas. Genetics. 1999; 152: 971-984.
- Saleh D, Xu P, Shen Y, Li C, Adreit H, Milazzo J, et al. Sex at the origin: an Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol Ecol. 2012; 21: 1330-1344.
- Tharreau D, Fudal I, Andriantsimialona D, Santoso, Utami D, Fournier E, et al. World population structure and migration of the rice blast fungus, Magnaporthe oryzae. Advances in Genetics, Genomics and Control of Rice Blast Disease. Springer Netherlands. 2009: 209-215.
- Zeigler RS. Recombination in Magnaporthe grisea. Annu Rev Phytopathol. 1998; 36: 249-275.
- Chuma I, Isobe C, Hotta Y, Ibaragi K, Futamata N, Kusaba M, et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011; 7: 2147.
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005; 434: 980-986.
- Hamer JE, Farrall L, Orbach MJ, Valent B, Chumley FG. Host species-specific conservation of a family of repeated DNA sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci USA. 1989; 86: 9981-9985.
- Kachroo P, Leong SA, Chattoo BB. Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea. Mol Gen Genet. 1994; 245: 339-348.
- Dobinson KF, Harris RE, Hamer JE. Grasshopper, a Long Terminal Repeat (LTR) retroelement in the phytopathogenic fungus Magnaporthe grisea. Mol Plant Microbe Interact. 1993; 6: 114-126.
- Wang Y, Kaye C, Bordat A, Adreit H, Millazzo J, Zheng X, et al. Construction of genetic linkage map and location of avirulence genes from cross CH63 and TH16 of Magnaporthe grisea. Zhongguo shuidao kexue. 2005; 19: 160-166.
- Zheng Y, Zhang G, Lin F, Wang Z, Jin G, Yang L, et al. Development of microsatellite markers and construction of genetic map in rice blast pathogen Magnaporthe grisea. Fungal Genet Biol. 2008; 45: 1340-1347.
- Dellaporta S, Wood J, Hicks J. A plant DNA minipreparation. Plant Mol Biol Rep. 1983; 1: 19-21.
- George ML, Nelson RJ, Zeigler RS, Leung H. Rapid population analysis of Magnaporthe grisea by using rep-PCR and endogenous repetitive DNA sequences. Phytopathology. 1998; 88: 223-229.
- Rolf F. Numerical taxonomy and multivariate analysis system ver. 2.02. Applied Biostatics Inc. 1998.
- Perrier X, Flori A, Bonnot F. Methods of data analysis. In: Genetic diversity of cultivated tropical plants. 2003: 33-63.
- Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst Biol. 2012; 61: 1061-1067.
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967; 27: 209-220.
- Seshu D, Kwak T, Mackill D. Global evaluation of rice varietal reactions to blast disease. 1986.
- Srinivasachary H, Shailaja, Shivayogi, Vaishali MG, Shashidhar HE, Kumar KG. Genetic analysis of rice blast fungus of southern Karnataka using DNA markers and reaction of popular rice genotypes. Curr Sci. 2002; 82: 732.
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980; 32: 314-331.
- Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, et al. Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Mol Breed. 1997; 3: 127-136.
- Nagaraju J, Reddy KD, Nagaraja G, Sethuraman B. Comparison of multilocus RFLPs and PCR-based marker systems for genetic analysis of the silkworm, Bombyx mori. Heredity. 2001; 86: 588-597.
- Consolo VF, Cordo CA, Salerno GL. Mating-type distribution and fertility status in Magnaporthe grisea populations from Argentina. Mycopathologia. 2005; 160: 285-290.
- Park SY, Milgroom MG, Han SS, Kang S, Lee YH. Genetic differentiation of Magnaporthe oryzae populations from scouting plots and commercial rice fields in Korea. Phytopathology. 2008; 98: 436-442.
- Takan JP, Chipili J, Muthumeenakshi S, Talbot NJ, Manyasa EO, Bandyopadhyay R, et al. Magnaporthe oryzae populations adapted to finger millet and rice exhibit distinctive patterns of genetic diversity, sexuality and host interaction. Mol Biotechnol. 2012; 50: 145-158.
- Notteghem J, Silue D. Distribution of the mating type alleles in Magnaporthe grisea populations pathogenic on rice. Phytopathology. 1992; 82: 421-424.
- Yaegashi H, Udagawa S. The taxonomical identity of the perfect state of Pyricularia grisea and its allies. Canadian Journal of Botany. 1978; 56: 180-183.
- Chen D, Zeigler RS, Leung H, Nelson RJ. Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology. 1995; 85: 1011-1020.
- Levy M, Correa-Victoria FJ, Zeigler RS, Xu S, Hamer JE. Genetic diversity of the rice blast fungus in a disease nursery in Colombia. Phytopathology. 1993; 83: 1427-1433.
- Levy M, Romao J, Marchetti MA, Hamer JE. DNA fingerprinting with a dispersed repeated sequence resolves pathotype diversity in the rice blast fungus. Plant Cell. 1991; 3: 95-102.
- Tosa Y, Hirata K, Tamba H, Nakagawa S, Chuma I, Isobe C, et al. Genetic constitution and Pathogenicity of Lolium isolates of Magnaporthe oryzae in comparison with host species-specific pathotypes of the blast fungus. Phytopathology. 2004; 94: 454-462.
- Farman ML, Eto Y, Nakao T, Tosa Y, Nakayashiki H, Mayama S, et al. Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol Plant-Microbe Interact. 2002; 15: 6-16.
- Rehmeyer C, Li W, Kusaba M, Kim YS, Brown D, Staben C, et al. Organization of chromosome ends in the rice blast fungus, Magnaporthe oryzae. Nucleic Acids Res. 2006; 34: 4685-4701.
- Chao C-CT, Ellingboe AH. Selection for mating competence in Magnaporthe grisea pathogenic to rice. Canadian journal of botany. 1991; 69: 2130-2134.
- Leslie JF, Klein KK. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics. 1996; 144: 557-567.
- Zeigler RS, Scott RP, Leung H, Bordeos AA, Kumar J, Nelson RJ. Evidence of parasexual exchange of DNA in the rice blast fungus challenges its exclusive clonality. Phytopathology. 1997; 87: 284-294.
- Gowda M, Shirke MD, Mahesh HB, Chandarana P, Rajamani A, Chattoo BB. Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genomics Data. 2015; 5: 284-291.
- Shirke MD, Mahesh H, Gowda M. Genome-Wide Comparison of Magnaporthe Species Reveals a Host-Specific Pattern of Secretory Proteins and Transposable Elements. PloS one. 2016; 11.
