Review Article
Austin J Pathol Lab Med. 2014;1(3): 5.
Long Non-Coding RNA MALAT1 Regulates Gene Expression and Protein Function via Multiple Layer and Flexible Manners
Xianyong Ma1*, Charles X Ma2, Cai Qiang3 and Xin Tang4
1Department of Pathology, Yale University School of Medicine, USA
2MD student, University of Connecticut School of Medicine, USA
3Department of Neurosurgery, Peoples Hospital of Wuhan University, China
4Department of Orthopedics Surgery, Peoples Hospital of Xiangxi, China
*Corresponding author: Xianyong Ma, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA
Received: October 15, 2014; Accepted: October 30, 2014; Published: November 03, 2014
Keywords
MALAT1; lncRNA; Gene expression and regulation; Mechanisms
Introduction
The Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) is one of the most significant molecules of long non-coding RNA (lncRNA), also known as Nuclear Enriched Transcript2 (NEAT2). MALAT1 was first discovered as a prognostic marker for non-small cell lung carcinoma [1]. Soon after, researchers found MALAT1 is linked to other cancers such as endometrial cancer [2], breast cancer [3], cervical cancer [4], colorectal cancer [5], hepatocellular carcinoma [6], liver cancer [7], neuroblastoma [8], osteosarcoma [9], pancreatic cancer [10], prostate cancer [11], bladder cancer [12], gastric cancer [13] and etc. In addition to its role as a biomarker for many human tumors, MALAT1 was also identified as a critical regulatory molecule to control target gene expression, modify RNA and protein (enzyme) activity, as well as affect cellular distribution [14-16], consequently it is intimately associated with the regulation of cell growth and proliferation. Dysregulations of MALAT1 result in multiple tissue carcinogenesis as well as many other human disease processes [17-20]. As such the critical regulatory role and potential clinical implications of MALAT1 have attracted more and more attention recently [10,17]. This review will focus on the regulatory role and molecular mechanisms of MALAT1 on gene expression and biochemical function of proteins. In addition, the paper will put forward the concept that the regulatory mechanisms of lncRNA on the gene expression and target protein function are in multiple layer and flexible manner.
I: MALAT1 regulates the bioactivity of target proteins via direct protein-lncRNA interaction
The interaction of lncRNA with protein has been linked to the regulation of target protein bioactivity [21-25]. MALAT1 interacts with several Alternative-Splicing (AS) factors such as SRSF1, SRSF2 and SRSF5 [26], which belong to a family of Serine/Arginine (SR)- rich splicing factors to regulate pre-RNA AS. Typically, SRSF proteins contain an RNA Binding Domain (RBD, also known as an RNA Recognition Motif, RRM) and Arginine/Serine (RS)-rich domain required for protein-protein interaction. SRSFs also regulate tissue or cell specific alternative splicing through a concentration and phosphorylation dependent manner. Full length MALAT1, as an abundant lncRNA molecule is localized in nuclear speckles and nucleoplasm, interacting with a sub-set of SRSF proteins and modulating their sub-nuclear distribution. It regulates cellular levels as well as the ratio of phosphorylated versus dephosphorylated SR proteins. Therefore MALAT1 plays a role in controlling alternative splicing patterns of certain endogenous pre-mRNAs. Ji Q. et al [27] demonstrated MALAT1 over expression in human Colorectal Cancer Cells (CRC), and discovered that MALAT1 binds to SFPQ (PTB-associated splicing factor), thus releasing proto-oncoprotein PTBP2 from SFPQ/PTBP2 complex, the increased SFPQ-detached PTBP2 then promoted cell proliferation and migration. The regulatory mechanism of protein activity by the interaction of MALAT1 and functional protein or complex is shown in Figure 1.
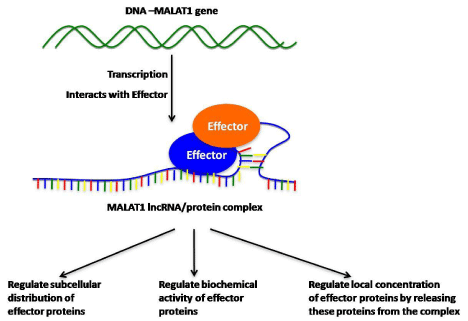
MALAT1 modifies the bioactivity of effector proteins.
MALAT1 molecule interacts with target proteins that contain RNA Binding Domain (RBD), such as alternative-splicing factors SRSF1, SRSF2, SRSF5 and SFPQ. It regulates the subcellular localization, enzymatic activity of these molecules and splicing behavior, and eventually affects the proliferation, differentiation and migration of cells.Figure 1:MALAT1 modifies the bioactivity of effector proteins.
MALAT1 molecule interacts with target proteins that contain RNA Binding Domain (RBD), such as alternative-splicing factors SRSF1, SRSF2, SRSF5 and SFPQ. It regulates the subcellular localization, enzymatic activity of these molecules and splicing behavior, and eventually affects the proliferation, differentiation and migration of cells.
II: MALAT1 regulates gene transcription via modification of epigenetic program
MALAT1 involvement in transcriptional regulations is also mediated through the epigenetic processes. Yang et al. [28] reports MALAT1 can facilitate the assembly of multiple co-repressors/co-activators and find that MALAT1 alters the histone modifications on chromatin by alternating the activity of Polycomb2 protein (Pc2) in vitro. For in vivo conditions, the interaction between MALAT1 and Pc2 protein causes to release the target genes from repressed status (in polycomb bodies) to activated form (in interchromatin granules) in response to stimulation of growth signals. In addition, the binding of unmethylated Pc2 proteins with MALAT1 lncRNA promotes E2F SUMOylation therefore activating the transcription processes for genes associated with growth control. Thus MALAT1 plays a regulation role in gene expression via modification of epigenetic programs. In addition, MALAT1 molecule has been linked to physically interact with critical chromatin-modifier Polycomb Repressive Complex 2 (PRC2) to modulate the epigenetic status of target genes. Sonia G. et al [29] reports MALAT1 directly binds to EZH2 protein, which is a critical component of PRC2 complex to play methyltransferase activity of the chromatin histone modifications; Jason AW. et al [30] publishes similar result that MALAT1 binds to active chromatin sites, these experimental evidences show MALAT1 modulates the chromatin histone methylations by binding to PRC2 complex and abolish the it’s methylation activity (Figure 2).
MALAT1 modifies the epigenetic program.
MALAT1 interacts with chromatin-modifying proteins such as Polycomb Repressive Complex (PRC2). MALAT1 releases the genes from repressed status (in polycomb bodies) to activation form (in interchromatin granules) by interaction with PRC2. It modulates the methyltransferase activity of histones and mediates the assembly of multiple coactivators and basal transcriptional machinery on the promoter region to initiate the gene transcription and alterative splicing.Figure 2:MALAT1 modifies the epigenetic program.
MALAT1 interacts with chromatin-modifying proteins such as Polycomb Repressive Complex (PRC2). MALAT1 releases the genes from repressed status (in polycomb bodies) to activation form (in interchromatin granules) by interaction with PRC2. It modulates the methyltransferase activity of histones and mediates the assembly of multiple coactivators and basal transcriptional machinery on the promoter region to initiate the gene transcription and alterative splicing.
III: MALAT1 affects the gene expression via chromosome translocation and gene fusion
Chromosome translocation causes changes in structure and conformation of chromosomes, frequently resulting in the production of new chimeric RNA and fusion proteins, which subsequently affects the gene transcription/translation activity and biochemical function of target proteins. It has been reported that MALAT1 gene is often translocated in tumor cells. Rajaram V. et al. [31] reports that t (11; 19) (q13.1; q13.42) is identified in Mesenchymal Hamartoma of Liver (MHL). This translocation causes a breakpoint in MALAT1 gene on chromosome 11, resulting in mutated isoforms of MALAT1 RNA by the fusion of MALAT1 sequence with the DNA from a gene-poor region termed MHLB1 on chromosome 19. The result is the production of mutated isoforms of MALAT1-MHLB1. This translocation and disruption of the MHLB1 region may contribute to MHL development through alterations in miRNA expression pattern. Renal cell carcinoma also often harbors MALAT1 translocations. Davis IJ. et al. [32] discovered t(6;11)(p21;q13.1) in a subset of renal cell carcinomas, this translocation causes MALAT1 (Alpha)- TFEB gene fusion, resulting in over-expression of native TFEB protein, which is not detectable under normal conditions. Therefore this translocation activates the TFEB expression or stabilizes the expression products (Figure 3).
MALAT1 affects the gene expression via chromosome translocations.
MALAT1 is located on chromosome 11, which is frequently rearrangement with other parts of chromosomes in tumor cells. (A). The t(6;11) (p21;q13.1) is found in a subset of human renal cell carcinomas, produces fusion transcript MALAT1-TFEB/TFEB-MALAT1, and leads to the up-regulation of wild type TFEB expression. (B). The t (11; 19) (q13.1; q13.42) is found in human mesenchymal hamartoma (MLHB region of chromosome) of the liver, resulting in production of abnormal chromosome 11 and 19 which alters miRNA expression pattern in chromosome 19. And this translocation may produce mutated MALAT1 transcripts, affecting the regulation function of MALAT1.Figure 3:MALAT1 affects the gene expression via chromosome translocations.
MALAT1 is located on chromosome 11, which is frequently rearrangement with other parts of chromosomes in tumor cells. (A). The t(6;11) (p21;q13.1) is found in a subset of human renal cell carcinomas, produces fusion transcript MALAT1-TFEB/TFEB-MALAT1, and leads to the up-regulation of wild type TFEB expression. (B). The t (11; 19) (q13.1; q13.42) is found in human mesenchymal hamartoma (MLHB region of chromosome) of the liver, resulting in production of abnormal chromosome 11 and 19 which alters miRNA expression pattern in chromosome 19. And this translocation may produce mutated MALAT1 transcripts, affecting the regulation function of MALAT1.
IV: Potential mechanisms of MALAT1 gene expression regulation
MALAT1 as a typical lncRNA molecule and potentially regulates gene expression via similar pathways that exist in other long non-coding RNAs such as HOTAIR (HOX transcript antisense RNA) [33], AIRN (antisense of IGF2R non-protein coding RNA) [34,35] and BANCR (BRAF-activated non-protein coding RNA) [36] and etc. Some critical mechanisms have already been identified which are involved in expression regulation by these lncRNA molecules.
- lncRNA interacts with gene promoters or enhancers to regulate transcription: The lncRNA-DNA-DNA triple helix was identified from lncRNA-chromatin interaction complex [37-40]. It has been reported that a GGUG-bearing lncRNAs (also called pncRNA, long promoter associated ncRNA) can recruit the RNA binding protein TLS (Translocation in Liposarcoma-associated) to the cyclin D1 promoter region and induce a conformational change of TLS. TLS then interacts with CBP/p300 and inhibits their HAT activities [41]. In this type of regulation, a key consensus sequence of the pncRNA is composed of GGUG, but not every lncRNA sequence containing GGUG was targeted by TLS, this suggests that a secondary structure of the GGUG-bearing lncRNAs is also involved in recognition by TLS. Furthermore there is no direct evidence to show the short fragments or full length MALAT1 bind to promoter DNA at present, it is nevertheless a reasonable prediction that MALAT1 may bind to some target promoter regions to regulate the transcription activity based on the fact that MALAT1 has plenty of GGUG conserved sequences (Figure 4a).
- lncRNA Interacts with mRNA as the antisense strand to block target translational activity: Recent researches [42,43] show the lncRNA H19 is processed into microRNA fragments (called miR675), which target tumor suppressor retinoblastoma (Rb) mRNA and down regulates its translation. Similarly, intronic antisense lncRNA fragments are correlated with the degree of tumor differentiation in prostate cancer samples [44]. MALAT1 transcripts display distinct subcellular localizations that may be associated with the unique functions. For example, a highly conserved 61 bp tRNA-like small RNA molecule from full length MALAT1 is identified exclusively in the cytoplasm [45], these small RNAs are named MALAT1-associated small cytoplasmic RNAs (mascRNA). Some of these mascRNAs of MALAT1 possibly act as siRNA molecules to bind to complementary coding mRNA sequences, which then down-regulate gene expression by degrading the target mRNA transcripts (Figure 4b).
- Regulation of gene expression via cis-splicing and trans-splicing processes: In eukaryotic cells, the primary transcripts include intronic and exonic sequences. The pre-mRNAs are processed by the spliceosome (splicing complex) to remove intronic sequences to form matured mRNA. When this process of intron removal occurs within the same pre-mRNA molecule, it is termed cis-RNA splicing. Splicing occurring in two separated pre-mRNA molecules is termed trans-RNA splicing. Both cis and trans RNA splicing processes follow similar mechanisms, which are catalyzed by the spliceosome [46]. Zhang, XO. et al [47] Recently reports sno-lncRNAs (intro-derived long noncoding RNAs with snoRNA [small nucleolar RNA] ends) are highly expressed from imprinted Prader-Willi Syndrome (PWS) region on human chromosome 15q11-q13. The sno-lncRNAs formation requires alternative splicing procedures, including cis and trans splicing. Thus a single gene can be spliced into multiple RNA transcripts. Using public database, the cis and trans variants of lncRNA including MALAT1 can be found [48,49], suggest similar mechanism may exist for MALAT1 functions (Figure 4c).
- lncRNA interacts with basal transcriptional machinery to regulate transcription activity: It has been discovered that lncRNAs can interact with basal transcription machinery of cell to influence transcription activity. For example, a 331 bp 7SK lncRNA represses transcription elongation via preventing pTEFß transcription factor from phosphorylating the RNAPII Carboxyl-Terminal Domain (CTD), therefore regulating the transcription activity [50]. Other authors [51] reports that a 178-nucleotide called B2 lncRNA plays a role via binding with RNAPII to inhibit phosphorylation of its CTD by TFIIH kinase activity, therefore abolishing the binding ability of RNAPII to promoter sequences and as a result repressing transcription (Figure 4d).
- lncRNA regulates gene expression and target protein activity through a multiple-layer and flexible manner: The regulatory manner of lncRNA shows a significant difference from the classic regulatory modality of protein factors.
- As lncRNA genes can easily produce different alternatively spliced isoforms from a single and fixed DNA sequence [47,52], it has been shown that the inflammatory signaling via TNF-α in mouse cells [53] can induce lncRNAs and its isoforms.
- lncRNA can change subcellular localization by migrating from nuclei to cytoplasm. For example, the full length MALAT1 is localized in nuclear speckles while small fragments derived from MALAT1 cleavage are localized in the cytoplasm. This kind of migration offers the ability of lncRNA molecule to interact not only with nuclear DNA or proteins, but also with cytoplasm mRNAs and protein factors [45,54], therefore, a single lncRNA molecule can play regulatory function at different stages of gene expression including transcription, translation and Posttranslational Modifications (PTM) of proteins. In addition to the regulations derived from interactions between lncRNA and target macromolecules, the regulations by lncRNAs are also affected by the interactions with the chemical environment including pH value, salt concentrations and other small molecules. All of these factors will affect the stability of and potential for such interactions.
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22: 8031-8041.
- Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006; 97: 106-112.
- Guffanti A, Iacono M, Pelucchi P, Kim N, Soldá G, Croft LJ, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009; 10: 163.
- Guo F, Li Y, Liu Y, Wang J, Li Y, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai). 2010; 42: 224-229.
- Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011; 39: 169-175.
- Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006; 44: 1012-1024.
- Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013; 26: 155-165.
- Koshimizu TA, Fujiwara Y, Sakai N, Shibata K, Tsuchiya H. Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci. 2010; 86: 455-460.
- Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010; 1799: 597-615.
- Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014; 15: 2971-2977.
- Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013; 190: 2278-2287.
- Zhang Q, Su M, Lu G, Wang J. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer. 2013; 12: 101.
- Wang J, Su L, Chen X, Li P, Cai Q, Yu B, et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014; 68: 557-564.
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013; 9: 1003368.
- Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014; 111: 736-748.
- Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73: 1180-1189.
- Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014; 55: 941-951.
- Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013; 97: 69-80.
- Watts R, Johnsen VL, Shearer J, Hittel DS. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013; 304: 995-1001.
- Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012; 488: 111-115.
- Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013; 34: 613-620.
- Gänther OP, Lin D, Balshaw RF, Ng RT, Hollander Z, Wilson-McManus J, et al. Effects of sample timing and treatment on gene expression in early acute renal allograft rejection. Transplantation. 2011; 91: 323-329.
- Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014; 204: 61-75.
- Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One. 2014; 9: e107016.
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014; 13: 92.
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010; 39: 925-938.
- Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014; 111: 736-748.
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011; 147: 773-788.
- Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012; 19: 664-670.
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014; 55: 791-802.
- Rajaram V, S Knezevich, Bove KE, Perry A, Pfeifer JD. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymalhamartoma of the liver harboring the t(11; 19)(q11; q13.4) translocation. Genes, chromosomes & cancer. 2007; 46: 508-513.
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003; 100: 6051-6056.
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007; 129: 1311-1323.
- Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, Stocsits RR, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012; 338: 1469-1472.
- Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, et al. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development. 2013; 140: 1184-1195.
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012; 22: 1006-1014.
- Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010; 24: 2264-2269.
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011; 44: 667-678.
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014; 514: 102-106.
- Kurokawa R. Promoter-associated long noncoding RNAs repress transcription through a RNA binding protein TLS. Adv Exp Med Biol. 2011; 722: 196-208.
- Oyoshi T, Kurokawa R. Structure of noncoding RNA is a determinant of function of RNA binding proteins in transcriptional regulation. Cell Biosci. 2012; 2: 1.
- Li H, Yu B, Li J, Su L, Yan M, Zhu Z, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014; 5: 2318-2329.
- Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010; 31: 350-358.
- Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004; 23: 6684-6692.
- Wilusz JE, Freier SM, Spector DL . 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008; 135: 919-932.
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003; 17: 419-437.
- Zhang XO, Yin QF, Wang HB, Zhang Y, Chen T, Zheng P, et al. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics. 2014; 15: 287.
- Frankish A, Mudge JM, Thomas M, Harrow J. The importance of identifying alternative splicing in vertebrate genome annotation. Database (Oxford). 2012.
- Chakraborty S, Deb A, Maji RK, Saha S, Ghosh Z. LncRBase: An Enriched Resource for lncRNA Information. PLoS One. 2014; 9: 108010.
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013; 11: 59.
- Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA represses TFIIH phosphorylation of RNA polymerase II. Transcription. 2011; 2: 45-49.
- Vitulo N, Forcato C, Carpinelli EC, Telatin A, Campagna D, D'Angelo M, et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014; 14: 99.
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013; 2: 00762.
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012; 18: 1487-1499.
- Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013; 190: 2278-2287.
- Lai MC, Z Yang, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Medical oncology (Northwood, London, England). 2012; 29: 1810-1816.
- Watts R, Ghozlan M, Hughey CC, Johnsen VL, Shearer J, Hittel DS. Myostatin inhibits proliferation and insulin-stimulated glucose uptake in mouse liver cells. Biochem Cell Biol. 2014; 92: 226-234.
Potential mechanisms of MALAT1 gene expression regulation.
Four different regulation mechanisms by lncRNAs are showed that are not identified in MALAT1 regulations, however, it is possible that these mechanisms also exist in MALAT1 regulations. (A). lncRNA molecule interacts with double strand DNA and represses gene transcription; (B). lncRNA fragments act as intronic siRNA to bind with mRNA and repressing mRNA translation; (C). Produce alternative splicing lncRNAs to regulate gene expression. Different isoforms from alternative splicing have different regulation activity and specificity, which regulate the gene expression with different patterns; (D). lncRNA molecule interacts with basal transcriptional machinery which disrupts the transcription initiation complex and represses transcription.Figure 4:Potential mechanisms of MALAT1 gene expression regulation.
Four different regulation mechanisms by lncRNAs are showed that are not identified in MALAT1 regulations, however, it is possible that these mechanisms also exist in MALAT1 regulations. (A). lncRNA molecule interacts with double strand DNA and represses gene transcription; (B). lncRNA fragments act as intronic siRNA to bind with mRNA and repressing mRNA translation; (C). Produce alternative splicing lncRNAs to regulate gene expression. Different isoforms from alternative splicing have different regulation activity and specificity, which regulate the gene expression with different patterns; (D). lncRNA molecule interacts with basal transcriptional machinery which disrupts the transcription initiation complex and represses transcription.
Gene regulations by lncRNA via flexible and multiple-layer manners signify the importance of physiological functions in efficiently balancing gene expression and biological behavior. Controlling or managing these regulations may also serve as new therapeutic strategies to different diseases including many types of cancers. The clinical implication and therapeutic potential of MALAT1 have attracted researchers to explore the down-regulation of this target and the results are encouraging. For example, Ren S. et al [55] reports in vitro and in vivo results of down-regulation for MALAT1 expression by siRNA technique, they find that for cultured prostate cells, down-regulation of MALAT1 inhibits prostate cancer cell growth, invasion and migration, as well as induces G0/G1 phase arrest. In vivo, intratumor delivery of therapeutic siRNA targets MALAT-1, delays tumor growth and reduces metastasis of cancer cell xenografts in castrated male nude mice. Surprisingly, this effect is consistent across different tumor types, Gutschner, TM. et al. [16] reports that targeted MALAT1 of lung cancer cells using siRNA inhibits metastasis formation after tumor implantation in vivo. Lai, MC. et al. [56] reports in a Hepatocellular Carcinoma (HCC) tumor cell line (HepG2), inhibition of MALAT1 could effectively reduce cell viability, motility, invasiveness, and increase the sensitivity to apoptosis. The clinical implications of MALAT1 have also been extended to other diseases. Watts, R. et al. [57] reports that knockdown of MALAT1 using siRNA suppresses myoblast proliferation by arresting cell growth in G0/G1 phase. Therefore target inhibition of MALAT1 shows potential for regenerative biology in the treatment of muscle atrophy and muscle wasting diseases. MALAT1 not only opens new avenues in considerations of gene expression regulation, it also potentially serves as one of the best predictive biomarker for many tumors. But perhaps most importantly, it provides a potential target for new and critical clinical therapeutics.
References