
Research Article
Austin J Pathol Lab Med. 2021; 8(2): 1033.
Association of Common Variant Rs2281135 of PNPLA3 with Alcohol-Related Cirrhosis in Chinese Han Males
Chen H1#, Yang F2#, Zhang Y1#, Zhang Y3, Guo T4, Mao Y5, Liu C6, Cheng L6, Wang Y7, Li Y6* and Huang J1*
1Department of Clinical Laboratory, The First Hospital of Jilin University, Changchun, China
2Department of Pediatrics Outpatient Service, The First Hospital of Jilin University, Changchun, China
3Department of Clinical Laboratory, Beijing Tiantan Hospital, Capital Medical University, Beijing, China
4Department of Clinical Laboratory, Beijing Mentougou District Hospital, Beijing, China
5Department of Center of Clinical Laboratory Medicine, The Fifth Medical Center of the General Hospital of the Chinese People’s Liberation Army, Beijing, China
6Department of Clinical Laboratory, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
7Department of Cancer Research Institute, The First Hospital of Jilin University, Changchun, China
#Co-First authors on this work
*Corresponding author: author: Jing Huang, Department of Clinical Laboratory, The First Hospital of Jilin University, Jilin, 130021, China
Yongzhe Li, Department of Clinical Laboratory, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100730, China
Received: April 27, 2021; Accepted: May 18, 2021; Published: May 25, 2021
Abstract
Background and Aim: Alcoholic Liver Disease (ALD), caused by longterm heavy alcohol consumption, is influenced by genetic factors. Studies have illustrated the overlapping genetic mechanism in Nonalcoholic Fatty Liver Disease (NAFLD) and ALD. Recently, a number of Genome-Wide Association Studies (GWAS) have demonstrated several SNPs were strongly associated with NAFLD. The aim of present study is to evaluate the association between these NAFLD-associated SNPs and ALD in Chinese Han population.
Methods: Nine SNPs were selected and genotyped in a cohort of 507 patients with ALD and 645 healthy controls by using MassARRAY iPLEX system. Alleles and genotypes analysis of SNPs were performed in logistic regression. The association between SNP and the level of liver serum biomarkers was tested in chi-square test and linear regression model.
Results: Our data confirmed that rs2281135 A-allele in PNPLA3 and rs3761472 G-allele in SAMM50 were significantly associated with increased risk of ALD (P = 1.93×10-12, OR [95% CI] = 1.82 [1.54-2.15]; P = 2.08×10-16, OR [95% CI] = 1.06 [1.04-1.08], respectively). The genotypes of rs2281135 were associated with ALD in additive, dominant and recessive genetic model (P = 1.24×10-11, P = 1.46×10-7, P = 2.07×10-9, respectively). In addition, rs2281135 was found to be associated with serum elevated levels of ALT (P = 5.0×10-3), AST (P = 0.03), ALP (P = 0.02), GGT (P = 0.03) in patients with ALD.
Conclusions: The present study confirmed that PNPLA3 common variant rs2281135 was significantly associated with ALD in Chinese male Han population.
Keywords: Alcohol-associated liver disease; Nonalcoholic fatty liver disease; Single nucleotide polymorphism; PNPLA3; SAMM50
Abbreviations
ALD: Alcoholic Liver Disease; AC: Alcoholic Cirrhosis; HCC: Hepatocellular Carcinoma; GWAS: Genome-Wide Association Study; SNP: Single-Nucleotide Polymorphism; PNPLA3: Patatin- Like Phospholipase Domain-Containing Protein 3; ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; GGT: Gamma- Glutamyl Transpeptidase; NAFLD: Non-Alcoholic Liver Disease; ALP: Alkaline Phosphatase; ALB: Albumin; HWE: Hardy-Weinberg Equilibrium
Introduction
Alcoholic Liver Disease (ALD) caused by long-term excessive alcohol consumption is a global public health issue and poses a significant burden on healthcare resources. ALD is a type of chronic liver disease that has a broad spectrum of progression, ranging from simple fatty liver to more severe forms of liver injury, including Alcoholic Hepatitis (AH), Alcoholic Cirrhosis (AC), even Hepatocellular Carcinoma (HCC) [1]. According to the Global Health Estimates (GHE) 2015 dataset by WHO, alcohol consumption was the second cause of deaths due to cirrhosis (20.8%) and liver cancer (29.8%) in the Asia-Pacific region, similar as in mainland China, 20.0% for all deaths due to cirrhosis and other chronic liver diseases and 32.5% for all deaths due to liver cancer, respectively [2]. The prevalence of ALD has increased rapidly in the developing countries, especially in China, but has decreased in Western countries [3].
The pathogenesis of ALD in humans is incompletely clear, genetic and environmental factors, including sex, ethnicity, obesity, drinking patterns and cigarette smoking et al. are associated with susceptibility to ALD [1]. Recently, a Genome-Wide Association Study (GWAS) [4] in population of European ancestry confirmed single-nucleotide polymorphism (SNP) rs738409 in patatin-like Phospholipase Domain-Containing Protein 3 (PNPLA3) was strongly associated with alcohol-associated cirrhosis (P = 1.54×10-48, OR [95%CI] = 2.19 [1.97- 2.43]), and identified Membrane-Bound O-Acyltransferase Domain- Containing Protein 7 (MBOAT7) gene polymorphisms (rs641738 and rs626283) and Transmembrane 6 Superfamily Member 2 (TM6SF2) gene polymorphisms (rs10401969 and rs58542926) as new risk loci for alcohol-associated cirrhosis. In our previous study, we tested the association between steatogenic genes (PNPLA3 rs738409, MBOAT7 rs626283 and rs641738, TM6SF2 rs58542926 and SUGP1 rs10401969) and ALD in Chinese Han population and validated that PNPLA3 rs738409 G-allele was associated with ALD (P = 1.25×10- 14, OR [95% CI] = 1.93[1.63-2.28]) and several biomarkers of liver including increased level of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl transpeptidase (GGT), and total bilirubin (TBil) in this cohort [5]. These SNPs also have been validated to be associated with non-alcoholic liver disease (NAFLD) [6-12], which demonstrated that ALD and NAFLD share overlapping pathogenic mechanism.
The study of NAFLD is faster than ALD, because of higher morbidity. GWAS is a powerful and broader approach to identify susceptibility genes for complicated human diseases. At present, a number of GWAS in NAFLD have been performed and identified multiple susceptibility loci associated with hepatic histology, increased level of serum AST and ALT or NAFLD activity score (NAS) in NAFLD [7,13-15]. Considering the overlap of genetic background in ALD and NAFLD and the association of these genetic variants with NAFLD, we hypothesized that these SNPs may be also associated with the prevalence of ALD in a Chinese Han population. Due to the different allele frequencies in Chinese Han population, we selected nine SNPs that Minor Allele Frequency (MAF) over 0.05 to investigate whether they were associated with ALD in Chinese Han population. Nine SNPs included rs2228603 in Neurocan (NCAN) gene, rs780094 in Glucokinase Regulatory Protein (GCKR) gene, rs2645424 in Farnesyl Diphosphate Farnesyl Transferase 1 (FDFT1) gene, rs1227756 in Collagen type XIII Alpha 1 (COL13A1) gene, rs6591182 in Latent Transforming Growth Factor-β Protein 3 (LTBP3) gene, rs6487679 in Pregnancy Zone Protein (PZP) gene, rs1421201 in solute carrier family 14 member 2 (SLC14A2), rs3761472 in sorting and assembly machinery component (SAMM50) gene and rs2281135 in PNPLA3 gene.
These SNPs have been verified to be associated with hepatic histology, increased level of serum AST and ALT or NAS in NAFLD. However, the role of these SNPs in development of ALD is currently unclear. The aim of this study was to test the potential association of nine variants with ALD in Chinese Han population.
Materials and Methods
Study population
We designed a multicenter case-control study, 507 individuals with alcohol-associated liver disease were collected from the first Hospital of Jilin University (Changchun, China), Peking Union Medical College Hospital (Beijing, China), the Fifth Medical Center of the General Hospital of the People’s Liberation Army (Beijing, China), Shengjing Hospital affiliated with China Medical University (Shenyang, China) and Hepatobiliary Hospital of Jilin (Changchun, China) from January 2016 to May 2017. All individuals were the outpatients or inpatients that diagnosed in five hospitals. The diagnostic criteria for ALD patients was based on the American Association for the Study of Liver Diseases (AASLD) practice guidelines (2010) [16], which revised by the Chinese Medical Association. The diagnosis of ALD was based on a combination of clinical features and characteristics, including history of alcohol consumption, ultrasonographic evidence of fatty liver or cirrhosis and supporting laboratory indicators abnormalities. Exclusion criteria for the study were as follows: 1) positivity to HBsAg, anti-HCV or HIV; 2) drug-induced liver disease, autoimmune liver disease, Wilson’s disease and other chronic liver disease. All subjects were unrelated Chinese Han male populations.
A total of 645 ethnicity-, age- and gender-matched healthy controls from the five hospitals were recruited during their routine physical examinations according to the following criteria: 1) had normal liver function, routine blood and urine results; 2) no history of HBV, HCV or HIV infection; 3) no evidence of liver disease in ultrasonography; and 4) no history of chronic disease.
All patients with ALD had a history of drinking for more than five years, the average alcohol intake is more than 40g per day for men or a large amount of alcohol consumption in the last two weeks averaging more than 80g per day. For healthy controls, the average alcohol consumption was less than 40g per day for men or did not had drinking habits in daily life. Clinical laboratory indicators were collected at baseline as following: ALT, AST, GGT, Alkaline Phosphatase (ALP), TBil, Albumin (ALB), Red Blood Cell Count (RBC), Mean Corpuscular Volume (MCV), Platelet (PLT) and Prothrombin Time (PT). The study was approved by The Ethics Committee of the First Hospital of Jilin University, and written informed consent was obtained from each subject.
Genotyping
2ml whole blood samples were collected into EDTA tubes from each individual by venous phlebotomy and stored at -80°C immediately after centrifuging. DNA was extracted from peripheral white blood cells of participants using Tiangen DNA extraction kit (Beijing, China) at the same time point, and then stored at -80°C until genotyped. Quality measurements were performed on all DNA samples. The nine SNPs (rs2228603, rs780094, rs2645424, rs1227756, rs6591182, rs6487679, rs1421201, rs3761472 and rs2281135) were genotyped using MassARRAY iPLEX system (Sequenom, USA). All the procedures were conducted following the manufacturer’s instructions. About 10-20ng DNA samples were amplified by Polymerase Chain Reaction (PCR) and the PCR products were performed to locus-specific single-base extension reactions. The final products were desalted and transferred to a 384-element SpectroCHIP array. Allele detection was carried out using matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOFMS). Then we used MassARRY Typer software v4.0 to analyze the mass spectrogram data.
Statistical analysis
To describe the study population, continuous traits were expressed as medians (and interquartile ranges) and categorical traits were expressed as numbers (and percentages). All statistical association analysis of genotype data was carried out using PLINK software v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/). To account for multiple testing, we used the False Discovery Rate (Bonferroni) correction for P value. The Hardy-Weinberg equilibrium (HWE) was estimated separately in the case group and the control group. If P≥0.05, genotype frequencies of the population followed the HWE.
Differences in baseline characteristics between ALD patients (n=507) and controls (n=645) was tested by Student’s t’-test or Mann-Whitney U test and variant genotype frequencies were tested by Chi-square test. To explore the association between genotypes and ALD, we performed three model analyses (additive model, dominant model, and recessive model) by logistic regression analysis. The associations between genotype and serum biomarkers of liver were tested by two models. Model 1 was performed by Chi-square test using the traditionally cutoff of over normal reference interval as elevation in ALD group. And model 2 was performed by linear regression to investigate the association of genotypes and liver serum biomarkers level in all participants. All serum markers measurements were natural logarithmically transformed before performing regressions, due to non-normal distributions.
All P-values (adjusted by Bonferroni correction) less than 0.05 was considered statistically significant and were adjusted for age in regression models. Linkage Disequilibrium (LD) test was carried out using Haploview software v4.2. Statistical analyses were carried out using SPSS Statistics V.25 (IBM) and Prism V.8 (GraphPad).
Results
Clinical characteristics of participants
The general characteristics of patients with ALD and controls are shown in Table 1. In present study, 507 ALD patients (median age 53 (47-59) years) and 645 healthy controls (median age 52 (45-59) years) were recruited from Chinese Han male population. All individuals were males, and age did not differ between two groups (P=0.21). Clinical characteristics comparisons between ALD group and control group, such as ALT, AST, ALP, GGT, ALB, RBC, MCV and PLT, were statistically significant (P<0.001).
Characteristics
ALD patients
Healthy Controls
P value
N
507
645
AC
472 (93.1%)
–
Age (years)
53 (47-59)
52 (45-59)
0.21
ALT (U/L)
23 (16-33)
23 (18-29)
***
AST (U/L)
41 (30-63)
20 (17-24)
***
ALP (U/L)
119 (90-163)
63 (53-75)
***
GGT (U/L)
63 (34-155)
26 (19-39)
***
ALB (g/L)
31 (27-35)
46 (45-48)
***
PLT (109/L)
78 (50-117)
224 (193-260)
***
RBC (1012/L)
3.30 (2.71-3.95)
4.93 (4.71-5.15)
***
MCV (fL)
96.3 (89.2-103.1)
90.3 (88.3-92.7)
***
Values are expressed as numbers (and percentages) for categorical traits, or median (and interquartile ranges) for continuous traits. P values were obtained by the Student’s t-test. AC: Alcoholic Cirrhosis; ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; GGT: Gamma-Glutamyl Transpeptidase; ALP: Alkaline Phosphatase; ALB: Albumin; PLT: Platelet; RBC: Red Blood Cell Count; MCV: Mean Corpuscular Volume.
***P value by Student’s t-test was less than 0.001.
Table 1: Baseline Characteristics of the Population.
We investigated 9 SNPs, of which SNP rs1421201 was excluded from further analysis, because the genotyping call rate (62.85%) of SNP rs1421201 in SLC14A2 gene was less than 80%. The SNPs details are shown in Table 2. The genotype of COL13A1 rs1227756 deviated from HWE (P = 0.01) in control population, whereas followed in ALD population (P=0.39). SAMM50 rs3761472 mutant G-allele was not found in control group, whereas rs3761472 genotype distribution followed Hardy-Weinberg Equilibrium (HWE) in ALD group (P>0.05). The genotype distributions of the remaining 6 SNPs followed HWE in the healthy controls (P >0.05).
SNP
Gene
CHR
Region
Allele
HWE test
rs2228603
NCAN
19
Missense variant
C>T
0.27
rs780094
GCKR
2
Intron variant
T>C
0.32
rs2645424
FDFT1
8
Intron variant
A>G
0.63
rs1227756
COL13A1
10
Intron variant
G>A
0.01a
rs6591182
EHBP1L1
11
Missense variant
T>G
0.12
rs6487679
PZP
12
Intergenic variant
C>T
0.88
rs1421201
SLC14A2
1
Intron variant
A>G
NA
rs3761472
SAMM50
22
Missense variant
A>G
NAa
rs2281135
PNPLA3
22
Intron variant
G>A
0.77
HWE: Hardy-Weinberg Equilibrium; NA: Not Available.
HWE test was performed on the genotype frequency distribution in the control population. If P=0.05, the genotype frequency of the population did not deviate from the HWE.
aThe genotype frequency in ALD population followed HWE.
Table 2: Nine SNPs information and HWE test.
Rs3761472 and rs2281135 were associated with ALD in the Han population
The genotyping information and genotype distributions of 8 SNPs in two groups are shown in Table 3 and Table 4, respectively. In logistic regression analyses with age as covariate (Table 3), we found that rs2281135 and rs3761472 were strongly associated with ALD. The mutant G-allele of rs3761472 was associated with an increased risk of ALD (OR [95% CI] = 1.06 [1.04-1.08], P=2.08×10-16). And the mutant A-allele of rs2281135 was also found to be associated with an increased risk of ALD (OR [95% CI] = 1.82 [1.54-2.15], P=1.93×10-12). As shown in Table 4, rs2281135 was associated with ALD in additive genetic model (OR [95% CI] = 1.78 [1.51-2.11], P=1.24×10-11), which illustrated that each copy of A-allele increased the risk of ALD about 1.78-fold. The genotypes of carrying of A-allele G/G& A/A and A/A were respectively also confirmed to increase the risk of ALD in dominant and recessive genetic model (Table 4). For rs3761472, A/G&G/G genotype showed the association with an increased risk of ALD in dominant genetic model (OR [95% CI] = 1.12 [1.09-1.16], P=8.83×10-18). Since no G-allele of rs3761472 was observed in the control group and no G/G genotype was observed in ALD group (81 individuals failed to be genotyped), the data of G/G genotype was not available. The analysis of additive and recessive genetic model could not be calculated. In addition, the genotype of rs3761472 deviated from HWE in control population, thus the effect of rs3761472 to increase ALD risk in Chinese Han population needed to be validated in bigger cohort.
Characteristics
ALD patients
Healthy Controls
P value
N
507
645
AC
472 (93.1%)
–
Age (years)
53 (47-59)
52 (45-59)
0.21
ALT (U/L)
23 (16-33)
23 (18-29)
***
AST (U/L)
41 (30-63)
20 (17-24)
***
ALP (U/L)
119 (90-163)
63 (53-75)
***
GGT (U/L)
63 (34-155)
26 (19-39)
***
ALB (g/L)
31 (27-35)
46 (45-48)
***
PLT (109/L)
78 (50-117)
224 (193-260)
***
RBC (1012/L)
3.30 (2.71-3.95)
4.93 (4.71-5.15)
***
MCV (fL)
96.3 (89.2-103.1)
90.3 (88.3-92.7)
***
Values are expressed as numbers (and percentages) for categorical traits, or median (and interquartile ranges) for continuous traits. P values were obtained by the Student’s t-test. AC: Alcoholic Cirrhosis; ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; GGT: Gamma-Glutamyl Transpeptidase; ALP: Alkaline Phosphatase; ALB: Albumin; PLT: Platelet; RBC: Red Blood Cell Count; MCV: Mean Corpuscular Volume.
***P value by Student’s t-test was less than 0.001.
Table 3: Association of allele in SNPs with ALD.
SNP
Model
Genotype
ALD
Control
OR (95% CI)
P value
rs2281135
Additive
G/G
120
246
1.78 (1.51-2.11)
1.24×10-11
G/A
235 (66.2)
300 (54.9)
A/A
151 (55.7)
96 (28.1)
Dominant
G/G
120 (24.0)
246 (38.3)
2.01 (1.55-2.60)
1.46×10-7
G/A & A/A
381 (76.0)
396 (61.7)
Recessive
G/G & G/A
355 (70.2)
546 (85.0)
2.42 (1.81-3.23)
2.07×10-9
A/A
151 (29.8)
96 (15.0)
rs3761472
Dominant
A/A
386 (88.9)
637 (100)
1.12 (1.09-1.16)
8.83×10-18
A/G & G/G
48 (11.1)
0 (0)
P values by logistic regression were adjusted for age (P value and OR of rs3761472 in general model were calculated by Chi-square test); NA: Not Available.
Table 4: Risk of ALD for genotypes in genetic models.
In our previous study, we demonstrated rs738409 of PNPLA3 G-allele was associated with an increased risk of ALD (P=1.25×10-14, OR [95% CI] = 1.93 [1.63-2.28]) in Chinese Han population5, which was performed in same cohort with this study. In this study, we confirmed rs2281135 of PNPLA3 A-allele was associated with an increased risk of ALD (P=1.93×10-12, OR [95% CI] = 1.82 [1.52- 2.15]) in Chinese Han population. Rs2281135 showed a consistency with rs738409, and these two SNP were both near the PNPLA3 gene. Therefore, the linkage disequilibrium was evaluated to determine whether rs2281135 was associated with rs738409. The two tag SNPs were in high linkage disequilibrium (D’ = 0.95, r² = 0.90) with each other.
The association between rs2281135 and biochemical markers of ALD
We verified the association between rs2281135 and biochemical markers of ALD in two models (Table 5). Rs2281135 was found to be associated with serum ALT (OR [95%CI] = 2.00 [1.22-3.27], P=5.0×10-3), AST (OR [95%CI] = 1.57 [1.01-2.39], P=0.03), ALP (OR [95%CI] = 1.67 [1.09-2.58], P=0.02) and GGT (OR [95%CI] = 1.61 [1.05-2.48], P=0.03) in model 1, but not with serum ALB (P>0.05). In model 2, we confirmed that the genotype of rs2281135 was associated with higher level of serum AST (P=0.02), ALP (P=2.0×10-3), lower level of serum ALB (P=8.82×10-11). However, we did not find the association between rs2281135 and serum ALT (P=0.17). As shown in Figure 1, the effect of PNPLA3 rs2281135 on serum levels of ALT, AST, ALP, GGT, and ALB was evaluated. The serum AST and ALP level of A/A genotype were 3.2U/L (relative change: 9.6%) and 10.1 U/L (relative change: 11.0%) higher than the G/G genotype, and 4.9g/L (relative change: 11.6%) lower than the G/G genotype.
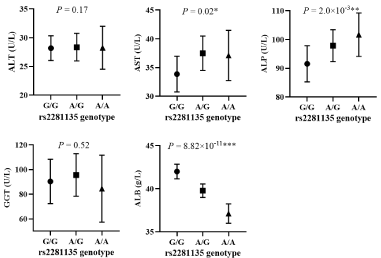
Figure 1: The association between rs2281135 and level of serum liver biomarkers. P was adjusted for age by linear regression in all participants. The frequencies of
G/G, A/G and A/A genotype in regression were 366, 535 and 247, respectively. ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; GGT: Gamma-
Glutamyl Transpeptidase; ALP: Alkaline Phosphatase; ALB: Albumin.
Liver marker
Model 1
Model 2
Comparisona
OR (95% CI)
P value
β (SE)
P value
ALT
P (n = 91) vs. N (n = 408)
2.00 (1.22-3.27)
-0.012 (0.009)
0.17
AST
P (n = 255) vs. N (n = 244)
1.57 (1.01-2.39)
0.03
0.024 (0.010)
0.02
ALP
P (n = 155) vs. N (n = 344)
1.67 (1.09-2.58)
0.02
0.027 (0.009)
GGT
P (n = 289) vs. N (n = 211)
1.61 (1.05-2.48)
0.03
0.011 (0.018)
0.52
ALB
P (n = 371) vs. N (n = 128)
0.70 (0.44-1.10)
0.12
-0.028 (0.004)
Model 1 was performed by Chi-square test in ALD group. Model 2 was adjusted for age by linear regression model in all participants. ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; GGT: Gamma-Glutamyl Transpeptidase; ALP: Alkaline Phosphatase; ALB: Albumin; aP: Patients positive for a certain phenotype; N: Patients negative for a certain phenotype.
Table 5: The association between rs2281135 and biochemical markers of ALD.
The regulatory effect of rs2281135
In order to illustrate the regulatory effect of intronic variant PNPLA3 rs2281135, we retrieved the HaploReg and RegulomeDB database. We found 21 SNPs in PNPLA3 gene were in high LD with rs2281135 by r² > 0.8 of LD threshold in Asian population. Among 21 SNPs, 14 SNPs with the rank <4 or the score >0.7, a variant (rs738409) and a variant (rs3747207) bounded with transcription factor were considered as available variant. Notably, rs738409, found in high LD with rs2281135 in Asian population, which was in active chromatin states and confirmed to have 2 transcription enhancer peaks (H3K4me1and H3K27ac) and 2 transcription promoter peaks (H3K1me3 and H3K9ac) by eQTL histone study, and changed 2 transcription factor binding motifs (WSR1-FL-1 and Pax_known3). Familiarly with rs738409, rs3747207 was also in active chromatin states and had 4 transcription enhancer/promoter peaks (H3K4me1, H3K27ac, H3K1me3 and H3K9ac), and changed a transcription factor binding motif (SIX5_known1). In addition, rs3747207 was demonstrated to have a protein bound with transcription factor GATA2 in SH-SYHY cell. For rs2281135, 2 transcription enhancer peaks (H3K4me1and H3K27ac) and a changed transcription factor binding motif were near rs2281135. In sum, intronic variant rs2281135, high LD with missense variant rs738409 that was verified to be associated with liver disease, may play an important role in progress of ALD accelerated by mutant of PNPLA3.
Discussion
ALD and NAFLD are the most prevalent causes of chronic liver injury in human liver disease and share common histological characteristics, progressing from hepatic steatosis to hepatitis, fibrosis and cirrhosis. The former occurs from habitually excessive alcohol intake, whereas the latter is closely related to metabolic syndrome such as type-2 diabetes mellitus or insulin resistance and abdominal obesity [17]. The occurrence and progression of these two diseases are both influenced by genetic and environmental factors. The susceptible loci that originally were identified to be associated with NAFLD and subsequently were confirmed to be associated with ALD [4,6,9,18,19], suggesting that genetic factors may affect the pathogenesis of these two types of chronic liver disease. In order to clarify this hypothesis, eight reported NAFLD-associated SNPs were genotyped in a cohort of 507 ALD patients and 645 healthy controls in Chinese Han population. In the present study, we confirmed the SNP rs2281135 in PNPLA3 gene and the SNP rs3761472 in SAMM50 gene were associated with the susceptibility of ALD in a Chinese Han population. Patients carrying A-allele of rs2281135 and G-allele of rs3761472 were at an increased risk of ALD pathogenesis.
The intronic variant rs2281135 at chromosome 22q13.31 of PNPLA3 gene was previously reported to be significantly associated with hepatic triglyceride content (P=6.9×10-12) in a multiethnic population-based exome-wide association study, which conferred susceptibility to NAFLD [7]. In a recent Korean population-based GWAS, Chung et al. demonstrated that rs2281135 showed a strong association with NAFLD (P=9.72×10-15) and severity of NAFLD (P=2.21×10-18) [20]. Our study demonstrated that PNPLA3 rs2281135 was associated with an increased risk of ALD in Chinese Han population. To explore the association between rs2281135 and serum level of liver biomarkers, we performed two models in different group. We found that the association of rs2281135 with elevated ALT was significant in ALD patients using the traditionally cutoff of over 40U/L indicating elevation. Other liver indicators were also associated with rs2281135 such as AST, ALP, GGT, but not ALB. In regression model, we failed to replicate the association between rs2281135 and level of serum ALT as previous study in Korean NAFLD population [20]. It may be because the disease cohort was different, and the ALT level deviation of the A/A genotype of rs2281135 population was large due to the small sample size. However, the level of AST, ALP and ALB was associated with rs2281135 in regression model.
PNPLA3, which was initially observed in adipose tissue by Baulande and coworkers [21], is a member of the patatin-like phospholipase family proteins and is predominantly expressed in adipose tissue. The missense variant rs738409 in PNPLA3 gene, which encodes an isoleucine to methionine substitution at 148, has shown a strong association with the major triglyceride hydrolase and adipose triglyceride lipase in adipose tissue [22,23]. Romeo et al. initially reported that rs738409 was associated with increased hepatic fat in Hispanic, African American and European populations [6]. Buch et al. demonstrated rs738409 as risk loci for alcohol-related cirrhosis (P=1.54×10-48) [4]. Subsequently other groups replicated the association between rs738409 and NAFLD [24-26] and ALD [5,27]. The current study confirmed that rs2281135 was in high linkage disequilibrium with rs738409 in this cohort. However, the function of intronic variant rs2281135 in PNPLA3 protein need to be clarified.
Rs3761472 of SAMM50 has been reported as a high susceptibility to the prevalence and progression of NAFLD in East Asian population [15,20,28]. The polymorphism of SAMM50 may be a key factor in mitochondrial dysfunction (partly due to the defective removal of reactive oxygen species). And several studies have demonstrated that mitochondrial dysfunction played a significant role in NAFLD [29,30] and ALD [31,32]. Our study conservatively revealed that carriers of the G-allele of rs3761472 were strongly associated with predisposition to ALD in Chinese Han population. However, no G-allele of rs3761472 was observed in the control group, which was inconsistent with previous NAFLD study by Chen et al. in Chinese Han population [15]. And no G/G genotype was observed in ALD group, thus the data in genetic model analysis was not available. More, the genotype of rs3761472 deviated from HWE in control group. Therefore, our data was insufficient to illustrate rs3761472 was associated with an increased risk of ALD in Chinese Han population, this effect needed to be validated in bigger cohort.
Yet there were several limitations in our study that should be mentioned. Firstly, 93.1% ALD patients was diagnosed as alcoholassociated cirrhosis, thus no stratified analysis in the ALD group. Secondly, because of traditional customs that females do not drink alcohol as frequently as males, our study excludes female ALD patients. And it is difficult to recruit enough available BMI in healthy control group. Therefore, only the age is used as co-variate in the multivariate analysis.
In summary, the present study first confirmed rs2281135 of PNPLA3 was associated with ALD in Chinese Han population. And the association between rs3761472 of SAMM50 and ALD in Chinese Han population needs to further verify in bigger cohort.
Disclosure Statement
Acknowledgements
We are most grateful to all participants in the present study. We also thank the colleagues from five hospitals who helped us collect the blood samples and clinical data.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the first Hospital of Jilin University. The medical records and biological specimens were obtained in previous clinical diagnosis and treatment; there is no need to contact the patient directly, which will not cause any damage to the participants.
References
- Gao B, Bataller R. Alcoholic Liver Disease: Pathogenesis and New Therapeutic Targets. Gastroenterology. 2011; 141: 1572-1585.
- WHO. Global Health Estimates 2015: deaths by cause, age, sex, by country and by region, 2000-2015. 2016.
- Liangpunsakul S, Haber P, McCaughan GW. Alcoholic Liver Disease in Asia, Europe, and North America. Gastroenterology. 2016; 150: 1786-1797.
- Buch S, Stickel F, Trepo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015; 47: 1443-1448.
- Zhang Y, Guo T, Yang F, et al. Single-nucleotide rs738409 polymorphisms in the PNPLA3 gene are strongly associated with alcoholic liver disease in Han Chinese males. Hepatol Int. 2018; 12: 429-437.
- Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008; 40: 1461- 1465.
- Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014; 46: 352-356.
- Lin YC, Chang PF, Chang MH, et al. Genetic variants in GCKR and PNPLA3 confer susceptibility to nonalcoholic fatty liver disease in obese individuals. Am J Clin Nutr. 2014; 99: 869-874.
- Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology. 2016; 150: 1219-1230.
- Goffredo M, Caprio S, Feldstein AE, et al. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: A multiethnic study. Hepatology. 2016; 63: 117-125.
- Krawczyk M, Rau M, Schattenberg JM, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res. 2017; 58: 247-255.
- Umano GR, Caprio S, Di Sessa A, et al. The rs626283 Variant in the MBOAT7 Gene is Associated with Insulin Resistance and Fatty Liver in Caucasian Obese Youth. Am J Gastroenterol. 2018; 113: 376-383.
- Chalasani N, Guo X, Loomba R, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 2010; 139: 1567-1576.
- Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011; 7: e1001324.
- Kitamoto T, Kitamoto A, Yoneda M, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013; 132: 783-792.
- O’Shea RS, Dasarathy S, McCullough AJ, et al. Alcoholic liver disease. Hepatology. 2010; 51: 307-328.
- Sanyal AJ, American Gastroenterological A. AGA technical review on nonalcoholic fatty liver disease. Gastroenterology. 2002; 123: 1705-1725.
- Dongiovanni P, Petta S, Maglio C, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015; 61: 506-514.
- Liu YL, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014; 5: 4309.
- Chung GE, Lee Y, Yim JY, et al. Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population. Gut Liver. 2018; 12: 316-323.
- Baulande S, Lasnier F, Lucas M, et al. Adiponutrin, a transmembrane protein corresponding to a novel dietary-and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001; 276: 33336-33344.
- Romeo S, Huang-Doran I, Baroni MG, et al. Unravelling the pathogenesis of fatty liver disease: patatin-like phospholipase domain-containing 3 protein. Curr Opin Lipidol. 2010; 21: 247-252.
- Zimmermann R, Strauss JG, Haemmerle G, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004; 306: 1383- 1386.
- Zhang L, You W, Zhang H, et al. PNPLA3 polymorphisms (rs738409) and non-alcoholic fatty liver disease risk and related phenotypes: a meta-analysis. J Gastroenterol Hepatol. 2015; 30: 821-829.
- Krawczyk M, Rau M, Schattenberg JM, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res. 2017; 58: 247-255.
- Petta S, Di Marco V, Pipitone RM, et al. Prevalence and severity of nonalcoholic fatty liver disease by transient elastography: Genetic and metabolic risk factors in a general population. Liver Int. 2018; 38: 2060-2068.
- Salameh H, Raff E, Erwin A, et al. PNPLA3 Gene Polymorphism Is Associated With Predisposition to and Severity of Alcoholic Liver Disease. Am J Gastroenterol. 2015; 110: 846-856.
- Chen L, Lin Z, Jiang M, et al. Genetic Variants in the SAMM50 Gene Create Susceptibility to Nonalcoholic Fatty Liver Disease in a Chinese Han Population. Hepat Mon. 2015; 15: e31076.
- Sunny NE, Bril F, Cusi K. Mitochondrial Adaptation in Nonalcoholic Fatty Liver Disease: Novel Mechanisms and Treatment Strategies. Trends Endocrinol Metab. 2017; 28: 250-260.
- Peng KY, Watt MJ, Rensen S, et al. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J Lipid Res. 2018; 59: 1977-1986.
- Nassir F, Ibdah JA. Role of mitochondria in alcoholic liver disease. World J Gastroenterol. 2014; 20: 2136-2142.
- Palma E, Riva A, Moreno C, et al. Perturbations in Mitochondrial Dynamics Are Closely Involved in the Progression of Alcoholic Liver Disease. Alcohol Clin Exp Res. 2020; 44: 856-865.