
Research Article
Austin J Pharmacol Ther. 2023; 11(1): 1170.
Small Molecule Inhibitors Targeting Endothelial IL-1β Receptor (IL-1R1): A Novel Approach to Atherosclerosis Therapy
Solanki K1#, Atre R1#, Sharma R1, Bezsonov E2,3,4, and Baig MS1*
1Department of Biosciences and Biomedical Engineering (BSBE), Indian Institute of Technology Indore (IITI), Simrol, Indore, 453552, India
2Laboratory of Angiopathology, Institute of General Pathology and Pathophysiology, 8 Baltiiskaya Street, 125315, Moscow, Russia
3Laboratory of Cellular and Molecular Pathology of Cardiovascular System, Avtsyn Research Institute of Human Morphology of Federal State Budgetary Scientific Institution “Petrovsky National Research Centre of Surgery”, 3 Tsyurupa Street, 117418, Moscow, Russia
4Department of Biology and General Genetics, I.M. Sechenov First Moscow State Medical University (Sechenov University), 8 Izmailovsky Boulevard, 105043, Moscow, Russia
#Equal Contribution.
*Corresponding author: Baig MSDepartment of Biosciences and Biomedical Engineering (BSBE), Indian Institute of Technology Indore (IITI), Simrol, Indore, 453552, India
Received: January 26, 2023; Accepted: March 09, 2023; Published: March 16, 2023
Abstract
The current study identified small molecule inhibitors targeting IL-1β receptor in inhibiting the atherosclerosis. IL-1β plays an important role in the progression of atherosclerosis by binding to the IL-1R1 receptor. IL-1β secreted by the macrophage induces the expression of cell adhesion molecules on the endothelial cell which causes recruitment of immune cells to the intimal site, promoting the atherosclerosis progression. It also releases IL-6 and MMPs which helps in thrombus formation. Thus, targeting IL-1β can help in limiting the recruitment of immune cell to the intimal site. IL-1RA is an antagonist which binds to the IL-1R1 receptor and limit its interaction with the IL-1β. Thus, critical residues on the IL-1R1 interacting with both the IL-1β and IL-1RA were analyzed. A total of 17 residues were found to be common between both the complexs in the 4A region. Further, in-vitro study suggests 12 critical residues on the IL-1R1 mediating the interaction with the IL-1β. Thus, small molecule inhibitors were used to target the critical residues on the IL-1R1 receptor binding with the IL-1β. Virtual screening in Schrodinger and Auto Dock Vina suggested six compounds interacting with most of the critical residues on the IL-1R1 receptor. Further, physicochemical and ADMET analysis of those compounds suggested good values in all the parameters. Thus, the screened compounds hold good potential to act as an anti-IL-1β inhibitor in limiting the progression of atherosclerosis. Further studies would be to check the efficacy and effectivity of the compounds at the in-vitro and in-vivo levels.
Keywords: IL-1β; IL-1R1; Small molecule inhibitor; Virtual screening; Atherosclerosis
Introduction
Atherosclerosis is chronic inflammatory disease in which plaque builds up in the wall of the arteries, leading to plaque formation. Rupture of this plaque leads to blood clot formation, and subsequently leads to sudden cardiac arrest. Thus, understanding the mechanism of plaque formation can help design a therapeutic strategy to limit its progression [1,2]. Macrophage plays an important role in foam cell formation and certain pro-inflammatory cytokines released by macrophage leads to further chronic consequences in development of plaque [3]. One such cytokines is IL-1β. IL-1β plays an important role in development of atherosclerosis. Uptake of Ox-LDL by macrophage leads to activation of signaling cascade, leading to IL-1β expression [4]. IL-1β induces expression of adhesion factors (such as ICAM-1 and VCAM-1) and chemokines (such as MCP-1), which helps in the adhesion and accumulation of inflammatory cells to the intimal site, initiating the plaque formation [5]. Cytokines such as IL-6 and MMPs are also induced by IL-1β. IL-6 helps in formation of thrombosis while MMPs (such as MMP-1, 8 and 13) ruptures the fibrous cap, leading to thrombus formation [6,7]. Recent study has also investigated the role of Nitric Oxide Synthase (NOS)-1 mediated expression of IL-1β in macrophage after Ox-LDL uptake and the expression of adhesion molecules on the endothelial cells [8,9]. Thus, therapeutic strategy targeting IL-1β would play an important role in decreasing the foam cell formation by the macrophage and in inhibition of atherosclerosis at every stage of the disease.
Several therapeutic strategies such as recombinant proteins, monoclonal antibodies and vaccines have been used to target the IL-1β signaling in atherosclerosis [4]. Anakinra, a recombinant human interleukin receptor antagonist which can antagonize IL-1β by binding to the IL-1β binding site on the IL-1R1 receptor limits the binding of the IL-1β to the receptor [10]. For instance, in phase 2 trial, anakinra was found to reduce the plaque by 30% in atherosclerotic mice, decrease triglyceride levels and macrophage infiltration in ApoE-/- mice [11]. But it requires regular injections which can led to adverse drug effects and block both IL-1a and IL-1β binding, which can be detrimental [12]. Hence, the non-specificity and case of daily administration limits its usage. Another approach is the usage of monoclonal antibodies. Canakinumab and Gevokizumab are the monoclonal antibodies which have been used to selectively target IL-1β by forming antigen-antibody complex and sequestering the IL1β binding to the receptor [13]. They have also shown prolonged half-life and doesn’t require daily administration [14]. But, during the in-vitro studies they have been found to be leading to the plaque instability in ApoE-/- mice [15]. Thus, alternative therapeutic strategies are required which provides higher specificity as well as doesn’t require daily administration is required to overcome the limitations of already known inhibitors.
Drug discovery is a time-consuming and expensive process. Structure Based Virtual Screening (SBVS) can help reduce the time and cost involved in researching new drugs. SBVS predicts the best interaction mode between the two molecules to form a stable complex, and uses scoring functions to estimate the non-covalent forces between the target and the ligand. The technique helps in prediction of the active lead molecules before their synthesis [16]. Other factors such as toxicity, bioavailability and efficacy of the compound can also be checked before moving to the in-vitro and in-vivo process, thus saving time and effort. The process starts with the identification of molecular target for a given compound followed by virtual screening to identify the active drug candidate. This is followed by lead optimization by improving the binding and physicochemical properties of the compound [16]. Many compounds have been in the market through virtual screening such as saquinavir, ritonavir and indinavir (treatment for HIV), dorzolamide (for glaucoma), boceprevir (for Hepatitis C), among others [16,17]. Thus, the current study undertook the approach of SBVS to identify the lead molecule(s) against the target receptor.
Thus, the current study deals with the identification of novel molecule inhibitors against IL-1β receptor (IL-1R1), leading to the blockage of signaling with the IL-1β. Critical residues on the IL-1β mediating the interaction with the receptor were identified and virtual screening was done to identify lead compounds blocking receptor-ligand interaction. Further, physicochemical and ADMET analysis of the compounds were done to check the pharmacokinetics and toxicity profile of the compounds. The screened compounds showed promising results in the in-silico study, but the in-vitro and in-vivo studies are required to check the efficacy and effectively of the compounds with the IL-1β receptor to limit the IL-1β signaling in atherosclerosis, leading to deceases in the atherosclerosis progression.
Methods
Analyses of the crystal structure of IL-1β and IL-1R1-Crystal structure of the IL-1β and IL-1R1 was retrieved from RCSB PDB (https://www.rcsb.org/) (PDB 4DEP). The structure consists of ternary complex of IL-1β-IL-1R1-IL-1RAcP. Single chain of IL-1β (Chain D) and IL-1R1 (Chain E) were considered for further study, deleting the other chains from the complex. The structures were further processed by deleting water molecules and heteroatoms and used for further study. Analysis of the crystal structure between IL-1RA-IL-1R1 was also done to compare the common interacting residues between the IL-1β and IL-1RA with the IL-1R1. Crystal structure of IL-1RA-IL-1R1 complex was downloaded from RCSB PDB (PDB 1IRA). The structure consists of single chain of IL-1RA (Chain A) and IL-1R1 (Chain B) interacting with each other. Water molecules and heteroatoms were deleted from the structure and used for further studies. 4A interacting residues between both the complexes were analyzed in UCSF Chimera [20] to check the difference in the binding of the ligand(s) with the domains of the IL-1R1 receptor and to decipher the common interacting residues between the complex.
Screening of small molecule inhibitor against IL-1R1 receptor- Interaction of IL-1β with the domain 1 and 2 of the IL-1R1 receptor is critical for its activity [18]. Hence, critical interacting residues in domain 1 and 2 of IL-1R1 with the IL-1β in the crystal structure were analyzed in UCSF Chimera [20]. Targeting these residues in the complex can help in the inhibition of the interaction of the receptor with the IL-1β. Hence, library of FDA-approved compounds was downloaded from ZINC-15 [22]. A total of 1614 compounds were retrieved from the database in SDF format. The resultant compounds were then converted to pdbqt by OpenBabel [29]. The ligands were then subsequently used for docking in Schrodinger [24] and AutoDockVina v1.2.0 [25,26]. Glide module of the Schrodinger suite was used for docking [23]. Crystal structure of the IL-1R1 (PDB 4DEP; Chain E) was retrieved from RCSB PDB (https://www.rcsb.org/). Protein preparation wizard of Schrodinger suite [30] was used to prepare the structure of the protein for docking. Structure was then refined and optimized in PROPKA at pH 7.0 [31] followed by minimization using OPLS4 force-field [32]. Receptor grid was then generated around the receptor covering the domain 1 and 2 of the receptor. LigPrep module of Schrodinger suite [33] was used to prepare ligands for docking in Schrodinger. For docking in AutoDockVina, polar hydrogen atoms and Kolmann charges were added on the crystal structure of IL-1R1 (PDB 4DEP; Chain E) in AutoDock 4 [34]. Grid box was then generated around the receptor covering the whole molecule (blind docking) and ligands were then docked with the receptor in AutoDockVina v1.2.0 [26]. Ligands were converted to pdbqt in OpenBabel [29] and used for docking in AutoDockVina. For short listing the compounds, the compounds positive in both the platforms were considered. Dock score up to -5 were considered for Schrodinger and its corresponding dock score up to -6 in AutoDockVina. Further, the compounds were selected based on its interaction with the 12 critical residues on the receptor. This was followed by ADMET analysis of the compounds to derive its pharmacokinetic properties.
ADMET analysis of the screened compounds- pkCSM server (https://biosig.lab.uq.edu.au/pkcsm/) [28] was used to analyze the physicochemical and ADMET properties of the compounds. pkCSM uses graph-based signatures of the compounds to analyze the ADMET properties of the compounds by comparing it with the available in-vitro data on the server. The compounds positive in the physiochemical and ADMET properties were finalized as a therapeutic molecule in inhibiting the IL-1β interaction with the receptor.
Result and Discussion
Analyses of the crystal structure of IL-1β and IL-1RAwith IL-1R1-IL-1β plays an important role as a pro-inflammatory cytokine in atherosclerosis. It is expressed as inactive pro-IL-1β form in macrophages stimulated by Ox-LDL, which later gets activated by various caspases. The active IL-1β can then bind to the IL-1R1 receptor and leads to its dimerization with IL-1RaP, further activating transcription factor NF-κB and contributes to the expression of pro-inflammatory cytokines [18]. IL-1β also leads to the expression of IL-6, ICAM-1, VCAM-1 and various MMPs (such as MMP-1, 8 and 13) which causes infiltration of various immune cells into the intima as well as leads to event of plaque instability and thrombosis [5,6]. Thus, it becomes evident to design a therapeutic strategy targeting the binding of the IL-1β with the receptor which can limit the chronic consequences of the atherosclerosis. IL-1RA is a natural antagonist present in the cellular system which binds to IL-1R1 receptor and inhibits the interaction of the receptor with the IL-1β, thus limiting the inflammatory signaling process [19]. Hence, it becomes important to study the common interacting residues between the IL-1β and IL-1RA with the IL-1R1 to design a therapeutic strategy against those residues.PDB structure of IL-1β and IL-1R1 was retrieved from RCSB PDB (PDB 4DEP) and 4A interacting residues between the complex were analyzed in UCSF Chimera [20]. To compare the interaction of the IL-1β with the antagonist IL-1RA, 4A residues between the IL-1RA and IL-1R1 complex (PDB 1IRA) were also analyzed in UCSF Chimera. Analysis of the interacting interface between the IL-1β and IL-1RA with the IL-1R1 receptor indicated that IL-1β interacted with most of the residues in the domain 1, 2 and 3 of the receptor, while IL-1RA interacted with domain 1 and 2 with minimal interaction in domain 3 of the receptor. Interaction of IL-1β with domain 3 of the receptor is important in mediating the function of the receptor. Binding of the IL-1β with the domain 3 of receptor changes the conformation of the receptor to a 20° from its actual orientation, which helps in the recruitment IL1-RAcP adaptor protein onto the receptor further activating the signaling cascade [21]. As IL-1RA interact minimally with the domain 3, it does not govern the change in the orientation of the receptor, hence signaling remains inactivated. Our approach was to target the domain 1 and 2 of the IL-1R1 receptor as there were maximum numbers of critical interacting residues between the IL-1β and IL-1RA with the IL-1R1 receptor in the 4A region. Thus, domain 1 and 2 on the receptor was used as a target to which both IL-1β and IL-1RA binds. Analyses of the 4A interacting residues between the IL-1β and IL-1RA in domain 1 and 2 of the receptor indicated significant similarity between the residues, with 17 residues found common in both the complex (Figure 1).

Figure 1: Crystal structure analyses of interaction of IL-1β and IL-RA with the IL-1R1 receptor: A.) Schematic representation of the interaction of IL-1β with the domain 1, 2 and 3 of the IL-1R1 (PDB 4DEP) and the 4A interacting residues of domain 1, 2 and 3 of the IL-1R1 with the IL1β. B.) Schematic representation of the interaction of IL-1RA with the IL-1R1 (PDB 4IRA) and the 4A interacting residues of domain 1, 2 and 3 of the IL-1R1 with the IL-1RA. Residues in bold indicates common residues between the complex while underlined residues are the critical residues in the domain 1 and 2 of IL-1R1 interacting with the IL-1β. IL1-R1 is highlighted as purple, IL-1β is highlighted as blue and IL-1RA is highlighted as orange. 4A interacting residues between the complex is highlighted as green.
In-vitro analysis have identified 12 critical residues on the domain 1 and 2 of IL-1R1 interacting with IL-1β [21]. Our analysis of 4A interacting residues also found 12 critical residues on the IL-1R1 interacting with the IL-1β (Figure 1). Thus, targeting these critical residues on the IL-1R1 can limit the binding of the IL-1β with the receptor.
Screening of small molecule inhibitor against IL1-R1: In-silico virtual screening approach was taken to screen small molecule inhibitors which can interact with the critical residues on the receptor, inhibiting its interaction with the IL- 1β. A total of 1614 FDA-approved compounds were downloaded from the ZINC15 library [22] and docked onto the IL-1R1 receptor in Schrodinger [23,24] and AutoDockVina v1.2.0 [25,26]. Compounds were short-listed based on its interaction with the critical residues on the domain 1 and 2 of the receptor with dock scores upto -5.0 for Schrodinger and its corresponding dock scores upto -6.0 in AutoDockVina. Out of 1614 compounds, six compounds were found to be interacting with at least 1 or more out of 12 critical residues on the receptor (Figure 2 & Table 1). Thus, these compounds were further used to derive the physicochemical and ADMET properties.
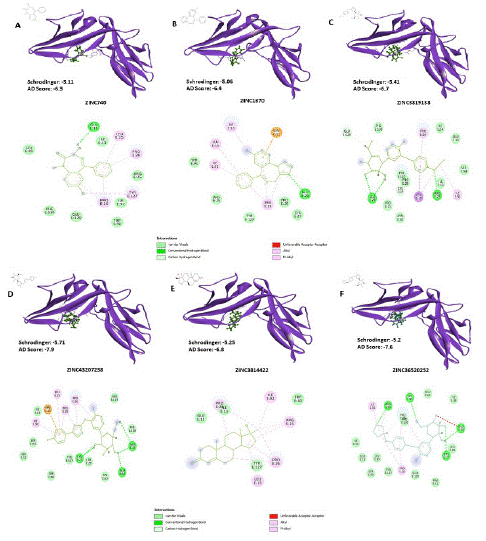
Figure 2: Docking of compounds with the IL-1R1 receptor. Schematic representation of docking of the compounds with the IL-1R1 receptor along with the 2D interaction diagram. A.) ZINC740, B.) ZINC1370, C.) ZINC3819138, D.) ZINC43207238, E.) ZINC3814422, and F.) ZINC36520252. Structures of the individual compounds are shown adjacent to the docked structures. Bonding interaction has been shown in different colour codes. IL-1R1 receptor is highlighted as purple while individual compounds have been highlighted as green.
Compounds
ZINC ID
Schrodinger
AutoDockVina
Dock Scores
Interacting residues*
Dock Scores
Interacting residues
Restoril
ZINC740
-5.11
111, 113, 124, 126, 127
-6.3
15, 127
Estazolam
ZINC1370
-5.06
111, 113, 124, 126, 127
-6.4
15, 127
Dapagliflozin
ZINC3819138
-5.41
14, 15, 127
-6.7
15, 127
Canagliflozin
ZINC43207238
-5.71
15, 127
-7.9
15, 127
Android
ZINC3814422
-5.25
15, 127
-6.8
15, 127
Empagliflozin
ZINC36520252
-5.2
15, 127
-7.6
15, 127
*Bold residue indicates H-bond
Table 1: Docking summary of the compounds interacting with the critical residues of IL-1R1.
Physicochemical and ADMET analysis of the top compounds: Analyzing the physicochemical and ADMET properties of the compounds can help in decreasing the risks during the clinical development, as it helps in evaluating the efficacy and safety of the drug during the development process. It also helps in limiting the failure of the drug during the pre-clinical and clinical trials [27]. Thus, in-silico approach was taken to determine the physicochemical and ADMET properties of the compounds using pkCSM server (https://biosig.lab.uq.edu.au/pkcsm/) [28]. pkCSM uses graph-based signature to determine the physicochemical and ADMET properties of the compound by comparing with the available database on the server. The result indicated that all six compounds were having positive value in terms of physiochemical and ADMET properties (Table 2). All
Parameters
ZINC740
ZINC1370
ZINC3819138
ZINC43207238
ZINC3814422
ZINC36520252
Physiochemical properties
M.W.
300.75
294.75
408.88
444.52
302.46
450.92
LogP
2.47
3.27
1.84
2.97
4.27
1.61
Rotatable bond
1
1
6
5
0
6
Acceptor
bond3
4
6
6
2
7
Donor bond
1
0
4
4
1
4
Absorption
Water Solubility
-3.607
-4.274
-4.346
-4.447
-4.33
-3.096
CaCo2 permeability
1.32
1.722
0.939
0.711
1.4
-0.035
Intestinal absorption
95.46
99.154
51.729
98.263
96.705
59.225
Skin permeability
-2.873
-2.438
-2.738
-2.735
-3.035
-2.765
Distribution
Fraction unbound
0.007
0.163
0.096
0.029
0.029
0.187
BBB permeability
0.316
0.506
-1.224
-1.186
0.184
-1.18
CNS permeability
-2.033
-1.425
-3.632
-3.303
-1.711
-3.7
Metabolism
CYP2D6 substrate
No
No
No
No
No
No
CYP2D6 inhibitor
No
No
No
No
No
No
Excretion
Total Clearance
0.21
0.266
0.198
0.039
0.625
0.402
Toxicity
AMES toxicity
No
No
No
No
No
No
Hepatotoxicity
No
No
No
No
No
No
Skin sensitization
No
No
No
No
No
No
Water solubility (logS)- defines solubility in water at 25°C; Skin permeability- logkp > - 2.5 classifies low skin permeability; Fraction unbound- defines unbound state in plasma protein remaining for pharmacological action; BBB permeability- logBB < - 1 classifies poorly distributed to the brain; CNS permeability- logPS > - 2 classifies CNS penetration and logPS < - 3 classifies no CNS penetration; Total clearance- includes both hepatic and renal clearance
Table 2: Physicochemical and ADMET properties of the selected compounds.
six compounds were following Lipinski rule of five. In absorption parameter, water solubility, Caco2 permeability, intestinal absorption and skin permeability were determined, in which all the parameters showed values above threshold. In distribution parameter, fraction unbound, blood-brain permeability and central nervous system permeability were determined. In metabolism, CYP2D6 substrate and CYP2D6 inhibitor were determined. In the excretion parameter, total clearance was determined, while in toxicity parameter, AMES toxicity, hepatotoxicity and skin sensitization were determined. Thus, the result suggests the effectivity of the screened compounds against inhibiting the IL-1β binding to the receptor.
Further studies would be to check these six compounds at the in-vitro and in-vivo level to confirm the efficacy of the inhibition of the IL-1β binding with the IL-1R1 receptor, which could lead to decrease in the atherosclerosis progression.
Conclusion
The study identified novel small molecule inhibitors targeting critical residues on the IL-1R1 receptor which helps in the binding of it with IL-1β. IL-1β plays an important role as a pro-inflammatory cytokine in progression of atherosclerosis and targeting its interaction with the IL-1R1 receptor can help in decreasing the atherosclerosis progression. The identified small-molecule inhibitors through in-silico screening holds good potential to work as an inhibitor of IL-1β, by analysis of its binding affinity as well as physicochemical and ADMET properties. Although, these in-silico studies have its limitation, in-vitro and in-vivo studies are required to validate these computational finding.
Consent for Publication
All the authors have consented for the publication in the CMLS.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Funding
This work was supported by Cumulative Professional Development Allowance (CPDA) from the Indian Institute of Technology Indore (IITI) to MSB.
Author Contributions
Conceptualization and investigation: MSB; Experiments: KS, RA, and RS; Writing (Original Draft): MSB, KS; Reviewing and editing MSB, EB, KS, RA, and RS. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
The authors acknowledge the Indian Institute of Technology Indore (IITI) for providing facilities and other support.
References
- Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014; 114: 1852-66.
- Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, et al. Atherosclerosis. Nat Rev Dis Primers. 2019; 5: 56.
- Moore KJ, FJ Sheedy, EA Fisher. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013; 13: 709-21.
- Mai W, Y Liao. Targeting IL-1beta in the Treatment of Atherosclerosis. Front Immunol. 2020; 11: 589654.
- Bevilacqua MP, Pober JS, Wheeler ME, Cotran RS, Gimbrone MA. Interleukin-1 activation of vascular endothelium. Effects on procoagulant activity and leukocyte adhesion. Am J Pathol. 1985; 121: 394-403.
- Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009; 27: 519-50.
- Loppnow H, P Libby. Proliferating or interleukin 1-activated human vascular smooth muscle cells secrete copious interleukin 6. J Clin Invest. 1990; 85: 731-8.
- Roy A, Saqib U, Wary K, Baig MS. Macrophage neuronal nitric oxide synthase (NOS1) controls the inflammatory response and foam cell formation in atherosclerosis. Int Immunopharmacol. 2020; 83: 106382.
- Roy A, U Saqib, MS Baig. NOS1-mediated macrophage and endothelial cell interaction in the progression of atherosclerosis. Cell Biol Int. 2021; 45: 1191-1201.
- Abbate A, Trankle CR, Buckley LF, Lipinski MJ, Appleton D, et al. Interleukin-1 Blockade Inhibits the Acute Inflammatory Response in Patients With ST-Segment-Elevation Myocardial Infarction. J Am Heart Assoc. 2020; 9: e014941.
- Ku EJ, Kim BR, Lee JI, Lee YK, Oh TJ, et al. The Anti-Atherosclerosis Effect of Anakinra, a Recombinant Human Interleukin-1 Receptor Antagonist, in Apolipoprotein E Knockout Mice. Int J Mol Sci. 2022; 23: 4906.
- Ramirez J, JD Canete. Anakinra for the treatment of rheumatoid arthritis: a safety evaluation. Expert Opin Drug Saf. 2018; 17: 727-732.
- Blech M, Peter D, Fischer P, Bauer MMT, Hafner M, et al. One target-two different binding modes: structural insights into gevokizumab and canakinumab interactions to interleukin-1beta. J Mol Biol. 2013; 425: 94-111.
- Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med. 2017; 377: 1119-1131.
- Gomez D, Baylis RA, Durgin BG, Newman AAC, Alencar GF, et al. Interleukin-1beta has atheroprotective effects in advanced atherosclerotic lesions of mice. Nat Med. 2018; 24: 1418-1429.
- Maia EHB, Assis LC, de Oliveira RA, da Silva AM, Taranto AG. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front Chem. 2020; 8: 343.
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014; 66: 334-95.
- Fields JK, S Gunther, EJ Sundberg, Structural Basis of IL-1 Family Cytokine Signaling. Front Immunol. 2019; 10: 1412.
- Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018; 281: 8-27.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004; 25: 1605-12.
- Krumm B, Y Xiang, J Deng. Structural biology of the IL-1 superfamily: key cytokines in the regulation of immune and inflammatory responses. Protein Sci. 2014; 23: 526-38.
- Sterling T, JJ Irwin. ZINC 15--Ligand Discovery for Everyone. J Chem Inf Model. 2015; 55: 2324-37.
- Friesner RA, Blanks JL, Murphy RB, Halgren TA, Klicic JJ, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004; 47: 1739-49.
- Schrodinger. Schrodinger Release 2021-2: Maestro v12.8. 2021: New York, NY.
- Eberhardt J, Martins DS, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J Chem Inf Model. 2021; 61: 3891-3898.
- Trott O, AJ Olson. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31: 455-61.
- Wu F, Zhou Y, Li L, Shen X, Chen G, et al. Computational Approaches in Preclinical Studies on Drug Discovery and Development. Front Chem. 2020; 8: 726.
- Pires DE, TL Blundell, DB Ascher. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J Med Chem. 2015; 58: 4066-72.
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, et al. Open Babel: An open chemical toolbox. J Cheminform. 2011; 3: 33.
- Schrodinger, Schrodinger Release 2021-2: Protein Preparation Wizard. 2021.
- Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput. 2011; 7: 525-37.
- Lu C, Wu C, Ghoreishi D, Chen W, Wang L, et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J Chem Theory Comput. 2021; 17: 4291-4300.
- Schrodinger, Schrodinger Release 2021-2: Ligprep. 2021: New York, NY.
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009; 30: 2785-91.