Review Article
Austin J Pharmacol Ther. 2014; 2 (2).1014
PRMT5, A Pivotal Player in Cancer
Rasika Mundade1, Han Wei1 and Tao Lu1,2,*
1Department of Pharmacology and Toxicology; Indiana University School of Medicine, USA
2Department of Biochemistry and Molecular Biology; Indiana University School of Medicine, USA
*Corresponding author: : Tao Lu, Department of Pharmacology and Toxicology; Indiana University School of Medicine, Indianapolis, USA
Received: February 03, 2014; Accepted: February 10, 2014; Published: February 13, 2014
Abstract
Protein arginine methyltransferases (PRMTs) are a family of enzymes that can add one or two methyl groups to the guanidino nitrogen atoms of arginine residues on histones and non–histone proteins. The abundant epigenetic modifications brought about by PRMTs help them regulate a wide variety of cellular functions, including RNA metabolism, transcriptional regulation, signal transduction, embryonic development and DNA damage repair, etc. Overexpression of different PRMTs has been frequently associated with many human cancers. Recently, increasing evidence suggests that PRMT5, an important member of the PRMT family, is a potential oncoprotein and is involved in tumorigenesis. Thus PRMT5 is an important target for therapeutic strategies. In this review, we present and discuss recent developments in our understanding of PRMT5 and its role in cancer.
Keywords Arginine, Cancer, Epigenetic regulation, Protein arginine methyltransferases
Abbreviations
aDMA– asymmetric dimethylarginine; AdoHcy– S–adenosyl homocysteine; AdoMet–S–adenosyl methionine; AKT–protein kinase B; AMI–1–protein arginine N–methyltransferase inhibitor 1; Ash2Labsent, small, or homeotic–like (Drosophila); CBP–CREB binding protein; CDK–cyclin–dependent kinase; CF Im68–cleavage factor Im68; COPR5–cooperator of PRMT5; CRAF–a member of the Raf kinase family of serine⁄threonine–specific protein kinases; CREBcAMP response element–binding protein; DNA–deoxyribonucleic acid; EBNA–Epstein–Barr virus nuclear antigen; EGFR–epidermal growth factor receptor; E2F–transcription factor in higher eukaryotes; FGFR–fibroblast growth factor receptor; HIF–hypoxia inducible factor; HoxA–homeobox protein A; JAK–Janus kinase; JBP–Janus kinase–binding protein; LSm–like Sm; MBP– myelin basic protein; Mep–methylosome protein; Men 1–multiple endocrine neoplasia type 1; MMA–monomethylarginine; NF–κB–nuclear factor κB; NM–nonmetastatic protein; PDCD–programmed cell death protein; p53–tumor suppressor 53; PI3K–phosphoinositide 3–kinase; PRC–polycomb repressive complex; PRMTs–protein arginine methyltransferases; R–arginine; Rad9–human homologue of cell cycle checkpoint control protein S. Pombe Rad9; Rb– retinoblastoma protein; RNA–ribonucleic acid; Saos–sarcoma osteogenic; sDMAsymmetric dimethylarginine; SHP–small heterodimer partner; Smsmall nuclear ribonucleoprotein–associated protein; snRNPs–small nuclear ribonucleoproteins; SPT–suppressor of Ty; ST–suppressor of tumorigenicity protein; SWI⁄SNF–SWItch⁄Sucrose nonfermentable; TIM–triosephosphate isomerase.
Introduction
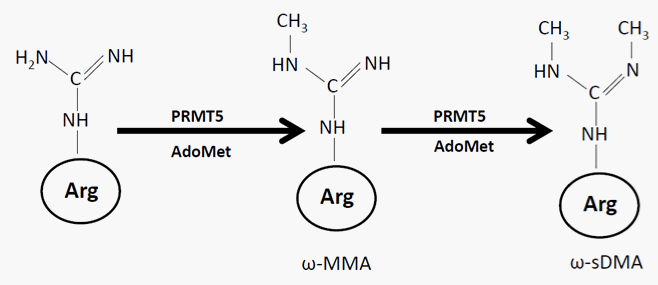
Arginine methylation is a common post–translational modification catalyzed by a family of intracellular enzymes termed protein arginine methyltransferases (PRMTs). PRMTs belong to the class of AdoMet (S–adenosyl–l–methionine) –dependent methyltransferases. PRMTs utilize AdoMet as a ubiquitous cofactor to catalyze highly specific methyl group transfers from methyl donor AdoMet, to the arginine residues on different biological targets. To date, ten PRMTs have been found in mammalian cells [1]. Based on the products of the enzymatic reactions, PRMTs can be classified as type I – IV enzymes. Type I enzymes catalyze the formation of ω–NG monomethylarginine (ωMMA) and asymmetric ω–NG, NG–dimethylarginine (ω–aDMA); Type II enzymes catalyze the formation of ωMMA and symmetric ω–NG, NG–dimethylarginine (ω–sDMA); Type III enzymes catalyze the formation of ωMMA only [2], and Type IV enzymes catalyze the formation of δ–NG–MMA [3]. PRMT5 is a typical type II methyltransferase (Figure 1). It is localized in both the nucleus and the cytoplasm and performs distinct functions by modifying either histones or non–histone proteins. For example, a study by Friesen WJ et al [4]demonstrated that, in the cytoplasm, PRMT5 is present in the ‘methylosome’ where it methylates Sm protein and such methylation is required for the assembly and biogenesis of snRNP core particles. Pal S et al [5]showed that, nuclear PRMT5 forms the complexes with the hSWI⁄SNF chromatin–remodeling proteins to methylate histone H3R8, therefore, decreasing the expression of tumor suppressor genes and acting as an oncogene. Fabbrizio’s group [6] identified a nuclear protein COPR5 (cooperator of PRMT5) which is required for nuclear functions of PRMT5. PRMT5 has also been shown to translocate from the nucleus to the cytoplasm at the time of extensive epigenetic reprogramming of mouse germ cells [7]. Study by Zhao Q et al [8] suggested that PRMT5 is predominantly localized in the nucleus in the bone marrow progenitors, whereas primarily localizedin the cytoplasm in the cord blood progenitors and may play a developmentally specific role in regulating gene expression at thehuman β–globin locus.
Figure 1: Chemical reactions catalyzed by PRMT5: PRMT5 is a typical type II enzyme. It utilizes AdoMet as methyl donor to add a single methyl on the terminal nitrogen atom of arginine to form ωMMA. It further adds an additional methyl group on the other terminal nitrogen, forming ω–sDMA. Abbreviations: AdoMet, S–adenosyl–l–methionine; ωMMA, ω – NG– monomethylarginine; ω–sDMA, symmetric ω – NG, NG– dimethylarginines.
Crystal structure of PRMT5
The crystal structure of full–length PRMT5 was initially determined by Sun et al from Caenorhabditis elegans [9]. This structure reveals that PRMT5 is composed of four domains: a TIM – barrel domain at the N–terminal end, a middle Rossmann–fold domain, a dimerization domain, and a C–terminal β–barrel domain [9]. The dimerization domain is inserted between β1 and β2 of the β–barrel domain [9]. The TIM – barrel, Rossmann–fold and β–barrel domains are packed together in a triangular manner with direct contacts between sequential domains [9]. In humans, PRMT5 functions as partof various high–molecular weight protein complexes that regulate its function and specificity [10]. These high–molecular weight complexes invariably contain the WD repeat–containing protein MEP50 (methylosome protein 50) [10]. Antonysamy et al (2012), reported the crystal structure of human PRMT5 in complex with MEP50 bound to an AdoMet analog and a peptide substrate derived from histone H4. The crystal structure of the hetero–octameric complex shows that the N–terminal domain of PRMT5 interacts very closely with the sevenbladed β–propeller MEP50, and delineates the structural elements of substrate recognition [10].
Role of PRMT5 in cancer signaling
PRMT5 was initially identified as Janus kinase (JAK) – binding protein 1 (JBP1). It can symmetrically methylate histones H2AR3, H3R2, H3R8 and H4R3 [1,11]. Recent studies reveal that PRMT5 can also methylate many non–histone proteins (Table 1), and many of these events are involved in tumorigenesis. For example, Pal et al (2004) showed that, in mouse embryonic fibroblast cell lines, PRMT5 acts as an oncogene by decreasing the expression of tumor suppressor genes like the suppressor of tumorigenicity 7 (ST7)and nonmetastatic 23 (NM23) [12]. Chung J et al [13] showed that inhibition of PRMT5 in non–Hodgkin lymphoma cell lines induceslymphoma cell death through reactivation of the Retinoblastoma (Rb) tumor suppressor pathway and Polycomb Repressor Complex 2 (PRC2) silencing, suggesting that inhibition of PRMT5 could be used as a promising therapeutic strategy for lymphoma. PRMT5 also plays a very important role in cell cycle progression and the DNA repair process. In human osteogenic sarcoma SAOS2 cells, PRMT5 increases sensitivity to DNA repair by methylating p53 at R333, R335, and R337 [14]. PRMT5 knockout in these cells induces the expression of p53 and causes p53–dependent apoptosis by triggering cell cycle arrest inthe G1phase, further confirming the role of PRMT5 in tumorigenesis in these cells. Moreover, PRMT5 has also been proven to function as an essential component of the hypoxia–inducible factor 1 (HIF–1) signaling pathway (15). HIF–1 is a key player in hypoxic response. A study by Lim et al [15] showed that, of the siRNAs targeting from PRMT1 to PRMT8, only the siRNA of PRMT5 attenuated the hypoxic induction of HIF–1a in human lung adenocarcinoma, fibrosarcoma, and mammary carcinoma cell lines, suggesting the uniquely important role of PRMT5 in these tumor cells. It is well known that both cyclin–dependent kinases (CDKs) and the phosphoinositide 3–kinase (PI3K)⁄AKT are the key players in cancer. PRMT5 is seen to upregulate CDKs and the PI3K⁄AKT signaling cascade, emphasizing its role as a potential oncoprotein [16]. Besides mouse and human cells, researchers also found that in C. elegans, PRMT5 methylates CREB–binding protein–1 (CBP–1) at R234 and leads to the inhibition of DNA damage–induced apoptosis [17]. Recently, our lab discovered that PRMT5 dimethylates R30 of the p65 subunit to activate the nuclear factor κB (NF–κB) [18]. Overexpression of PRMT5 increases NF–κB activity, while knockdown of PRMT5 greatly reduces NF–κB transactivation. Since NF–κB is a family of transcription factors that regulate a variety of cellular processes and its aberrant activation is frequently seen in diverse human cancers, data from our lab strongly suggest that PRMT5 is a tumor promoter possibly through the activation of NF–κB signaling [18].
Table 1: Known non–histone protein substrates of PRMT5
In addition to in vitro cell systems, the importance of PRMT5 in cancer has recently been realized in in vivo models and patient samples. Gu et al showed that knocking down PRMT5 in lung adenocarcinoma A549 cells partially downregulates the fibroblast growth factor receptor signaling pathway (FGFR), which reduces the cell growth and tumor xenografts in nude mice [19]. In an orthotopic model of breast cancer, Powers et al showed that tumor suppressor programmed cell death 4 (PDCD4) is methylated by PRMT5 at R110 [20]. Also, Ash2L (absent, small, or homeotic) like (Drosophila), a componentof mammalian histone H3K4 methyltransferase complexes associated with the transformation of human tumors, is methylated by PRMT5 at R296 [21]. A study by Cho et al [22] further demonstrated thatarginine methylation controls growth regulation by E2F–1. Analysisof a subgroup of colorectal cells showed that high levels of PRMT5coincided with low levels of E2F–1 and poor prognosis [22]. As PRMT5 was initially identified as JBP1, it is striking to see that most patients with myeloproliferative neoplasms express a constitutivelyactivated form of JAK2: JAK2–V617F [23]. This JAK2 mutant interactswith PRMT5 more strongly than the wild–type form, downregulates its methyltransferase activity, and promotes myeloproliferation [23].Recently, an interesting study by Hua’s group [24] in mice with excised Men1 gene (multiple endocrine neoplasia type 1) demonstrated that PRMT5 can interact with menin and suppress the Hedgehog signaling by dimethylating histone H4R3. This further potentiates PRMT5 as a therapeutic target in treating MEN1 tumors. Very recently, our lab reported findings from Oncomine data suggesting that PRMT5 ishighly overexpressed in many human cancers such as liver, pancreas,skin, breast, cervix, prostate, kidney, ovary, bladder, and lung, with strikingly elevated expression in colon cancer [18]. PRMT5 is a house keeping gene and complete loss of PRMT5 enzyme is not compatiblewith mouse or cell viability. Surani’s group [25] examined PRMT5 knockout blastocysts cultured and demonstrated that PRMT5 is required for embryonic epiblast cell differentiation and loss of PRMT5 leads to early embryonic lethality of mice.
Perspective: PRMT5 as a cancer therapeutic target
Since over–expression of PRMTs is associated with many human cancers, this makes PRMTs an ideal prognostic biomarker and potential therapeutic target for cancer treatment. To date, there are very few PRMT inhibitors that have been identified. For example S–adenosyl homocysteine (AdoHyc) is an isotype nonspecific PRMT inhibitor, which competes with products of methyltransferase reactionto bind to the active site of the enzyme [26]. Recently, small molecules have been identified that are specific to PRMTs with high potency invitro. AMI–1 (protein arginine N–methyltransferases inhibitor 1) is a small molecule inhibitor which inhibits the methyltransferase activity of only arginine but not lysine without competing for the AdoMet binding site [27]. Another small molecule inhibitor which has beensynthesized and acts as an irreversible inhibitor of PRMT1 is C21. C21 is a chloroacetamidine bearing histone H4 tail analog [28]. Somepyrazole–based highly selective and potent inhibitors for PRMT4 havealso been identified. Baiocchi’s group recently identified CPD5 as a novel specific inhibitor of PRMT5. CPD5 can inhibit proliferation and induce cell cycle regulation and apoptosis in lung cancer model [29]. Very recently, Folk et al identified small molecule inhibitors of PRMT5 with 100–fold greater inhibition of PRMT5 methylation and improved physicochemical properties. They used robust non–radiometric assay of peptide substrate methylation based on Transcreener EPIGEN technology and the compounds identified display on–target activity in cell lines and reduce proliferation of cancer cells [30]. Like other therapeutic targets in cancer, targeting PRMT5 will surely have its own side effect. However, this could only be further tested in clinical trials. Given the especially important role of PRMT5 in cancer, it is not surprising that more inhibitors will be identified for this important enzyme. As we described above, the crystal structure of human PRMT5:MEP50 complex has been recently reported (10). This information is of extreme importance, as it will surely accelerate the progress toward the synthesis of novel inhibitors of PRMT5 and treatment of cancer.
Acknowledgements
We thank Ms. Lisa King at the Department of Pharmacology and Toxicology at Indiana University School of Medicine for her generous and professional help with revising this review. The research is supported by grants 23–862–07TL (to TL) and 036433730102 (to TL).
References
- Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007; 120:4243- 4246.
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013; 13: 37-50.
- Fisk JC, Read LK. Protein arginine methylation in parasitic protozoa. Eukaryot Cell. 2011; 10: 1013-1022.
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modiied Sm proteins. Mol Cell Biol. 2001; 21: 8289-8300.
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/ SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004; 24: 9630-9645.
- Lacroix M, El Messaoudi S, Rodier G, Le Cam A, Sardet C, Fabbrizio E. et al. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008; 9: 452-458.
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006; 8: 623-630.
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009; 16: 304-311.
- Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, et al. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci U S A. 2011; 108: 20538-20543.
- Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, et al. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A. 2012; 109: 17960-17965.
- Wei H, Mundade R, Lange KC, Lu T. Protein arginine methylation of non- histone proteins and its role in diseases. Cell Cycle. 2013; 13:14-23.
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/ SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004; 24: 9630-9645.
- Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW, et al. Protein Arginine Methyltransferase 5 (PRMT5) Inhibition Induces Lymphoma Cell Death through Reactivation of the Retinoblastoma Tumor Suppressor Pathway and Polycomb Repressor Complex 2 (PRC2) Silencing. J Biol Chem. 2013; 288:35534-35547.
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008; 10: 1431-1439.
- Lim JH, Choi YJ, Cho CH, Park JW. Protein arginine methyltransferase 5 is an essential component of the hypoxia-inducible factor 1 signaling pathway. Biochem Biophys Res Commun. 2012; 418: 254-259.
- Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/ cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012; 103: 1640-1650.
- Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C, et al. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009; 5:e1000514.
- Wei H, Wang B, Miyagi M, She Y, Gopalan B, Huang DB, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-?B. Proc Natl Acad Sci U S A. 2013; 110: 13516-13521.
- Gu Z, Gao S, Zhang F, Wang Z, Ma W, Davis RE, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012; 446: 235-241.
- Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011; 71: 5579- 5587.
- Butler JS, Zurita-Lopez CI, Clarke SG, Bedford MT, Dent SY. Protein-arginine methyltransferase 1 (PRMT1) methylates Ash2L, a shared component of mammalian histone H3K4 methyltransferase complexes. J Biol Chem. 2011; 286: 12234-12244.
- Cho EC, Zheng S, Munro S, Liu G, Carr SM, Moehlenbrink J, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012; 31: 1785- 1797.
- Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, et al. JAK2V617F- mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011; 19: 283-294.
- Gurung B, Feng Z, Iwamoto DV, Thiel A, Jin G, Fan CM, et al. Menin epigenetically represses Hedgehog signaling in MEN1 tumor syndrome. Cancer Res. 2013; 73: 2650-2658.
- Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2011; 24: 2772-2777.
- Kuhn P, Xu W. Protein arginine methyltransferases: nuclear receptor coregulators and beyond. Prog Mol Biol Transl Sci. 2009; 87: 299-342.
- Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT, et al. Small molecule regulators of protein arginine methyltransferases. J Biol Chem. 2004; 279: 23892-23899.
- Obianyo O, Causey CP, Jones JE, Thompson PR. Activity-based protein proiling of protein arginine methyltransferase 1. ACS Chem Biol. 2011; 6: 1127-1135.
- Otterson GA, Wu X, Welliver M, Sharma S, Shilo K, Hitchcock C, et al. PRMT5, a Novel Epigenetic Target in Lung Cancers. Eur J Cancer 2012; 171.
- Falk H, Allan E, Yang H, Lowes K, Bergman Y, Walker S, Kersten W, et al. Development of Arginine Methyltransferase PRMT5 Inhibitors as Novel Agents for the Treatment of Cancers. CANCER 2013; 25th Lorne Cancer Conference 2013.
- Baldwin GS, Carnegie PR. Speciic enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971; 171: 579-581.
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. et al. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000; 275: 17221- 17129.
- Barth S, Liss M, Voss MD, Dobner T, Fischer U, Meister G, et al. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine-glycine repeat to the survival motor neuron protein. J Virol. 2003; 77: 5008-5013.
- Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003; 11: 1055-1066.
- Shire K, Kapoor P, Jiang K, Hing MN, Sivachandran N, Nguyen T, Frappier L. Regulation of the EBNA1 Epstein–Barr virus protein by serine phosphorylation and arginine methylation. J Virol 2006; 80:5261-5272.
- Martin G, Ostareck-Lederer A, Chari A, Neuenkirchen N, Dettwiler S, Blank D, et al. Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA. 2010; 16: 1646-1659.
- Andreu-Pérez P, Esteve-Puig R, de Torre-Minguela C, López-Fauqued M, Bech-Serra JJ, Tenbaum S, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011; 4:ra58.
- Hsu JM, Chen CT, Chou CK, Kuo HP, Li LY, Lin CY, et al. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011; 13: 174-181.
- He W, Ma X, Yang X, Zhao Y, Qiu J, Hang H, et al. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 2011; 39: 4719-4727.
- Kanamaluru D, Xiao Z, Fang S, Choi SE, Kim DH, Veenstra TD, et al. Arginine methylation by PRMT5 at a naturally occurring mutation site is critical for liver metabolic regulation by small heterodimer partner. Mol Cell Biol. 2011; 31: 1540-1550.
- Bandyopadhyay S, Harris DP, Adams GN, Lause GE, McHugh A, Tillmaand EG, et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012; 32: 1202- 1213.