Research Article
Austin J Pharmacol Ther. 2014; 2 (3). 1017
RAD51 135G>C Polymorphism and Cancer Risk: An Updated Meta–Analysis Involving 54,239 Subjects
Gui-li Sun1†*, Bei-Bei Zhang2†, Chao Xuan3, Kai- Feng Deng4, Ge Gao5,Li-Min Lun3
1Department of Endocrinology, The Second Hospital of Nanning City, The Third Affiliated Hospital of Guangxi MedicalUniversity, Nanning 530000, PR China
2Graduate School of Medicine, Mie University, Mie, Japan
3Department of Clinical Laboratory, The Affiliated Hospital of Medical College, Qingdao University, Qingdao, PR China
4Department of Clinical Laboratory, The Fourth Affiliated Hospital of Guangxi Medical University, Liuzhou, PR China
5Center for Reproductive Medicine,Tianjin Central Hospital of Obstetrics and Gynecology,Tianjin,PR China † Contribute equally to this work;
*Corresponding author: : Gui-li Sun, MD, Department of Endocrinology, The Second Hospital of Nanning City,The Third Affiliated Hospital of Guangxi MedicalUniversity, Nanning, PR China
Received: February 05, 2014; Accepted: March 28, 2014; Published: April 07, 2014
Abstract
The RAD51 plays a pivotal role in homologous recombination repair of DNA double-strand breaks inducing chromosomal breaks and genomic instability. Previous studies yielded conflicting results for the association between RAD51 135G>C polymorphism and risk of cancer. The present study aimed at investigating the pooled association using a meta-analysis on the published studies, involving 27,895 cases and 26,344 controls to assess the effect of RAD51 135G>C on cancer susceptibility. Across all populations, our results indicated that significant associations were found between RAD51 135G>C polymorphism and risk of cancer under genotypic C allele vs. G allele (OR =1.36 95% CI: 1.31–1.41), CC vs. GG (OR =2.37 95% CI: 2.12–2.65), CC vs. CG (OR =4.02 95% CI: 3.62–4.46), recessive model (OR =3.74, 95% CI: 3.40–4.11), and dominant model (OR =1.08, 95% CI: 1.03–1.13). In subgroup analyses, similar associations were found among Caucasians but not Asians. Moreover, the significant associations were found in subgroups of breast cancer, hematologic malignances, colorectal cancer, endometrial cancer, and ovarian cancer. This meta–analysis suggests that the RAD51 135G>C polymorphism was associated with susceptibility of cancer. The effect of the variants on the expression levels and the possible functional role of the variants in cancer should be addressed in further studies.
Key words: RAD51, Polymorphism, Cancer risk, Meta–analysis
Introduction
Epidemiologic studies reveal a significant environmental contribution to the pathogenesis of cancer [1,2]. Familial aggregation and twin studies indicate that the presence of genetic factors are for susceptibility to this condition [3–5]. A number of genomic screens have been performed to find genetic linkage to cancer [6–8]. The faithful repair of DNA damage such as chromosomal double–strand breaks (DSBs) is crucial for genomic integrity [9]. DSBs may cause chromosomal breaks and genomic instability, thus increasing the probability of developing cancer [10]. Homologous recombination (HR), single–strand annealing and non–homologous end–joining are considered to be the main pathways for repairing the DSBs [11]. Among them, the central HR protein is RAD51 which ensures high fidelity DNA repair by facilitating strand exchange between amaged and undamaged homologous DNA segments [10]. Thus far, two SNPs (135G⁄C [rs1801320] and 172G⁄T [rs1801321]) were discovered in the 5 UTR of RAD51 [12]. The effect of 135G→C variant on the RAD51 was alternative splicing within the 5 UTR, while the latter SNP was found to have weak effect [13].
The genetic variations of RAD51 gene may contribute to the development and progression of cancers [14]. Many original studies have reported the role of RAD51 135G>C polymorphism and cancer risk, but the findings are inconclusive [15,16]. Partially, it may due to the fact that the RAD51 gene was a minor gene for risk of cancers and⁄or the relatively small sample–size in each published studies.Therefore, we performed this updated meta–analysis to derive a more precise estimation of the association between RAD51 135G>C polymorphism and cancer.
Materials and methods
Selection of published studies
Case–control studies reporting the association between the RAD51 135G>Cpolymorphisms and risk of cancer published in English before February 2013 were identified by comprehensive computer–based searches of Medline, EBSCO, and BIOSIS databases. The references of reviews and retrieved articles were also searched simultaneously to find additional eligible studies. The following keywords were used for searching: “RAD51” AND (“genetic variant*” or “genetic variation” or “polymorphism*”) AND (“cancer” or “carcinoma” or “tumor” or “leukemia” or “laukaemia”). The most complete and recent results were used when there were multiple publications from the same study group.
Two investigators reviewed all identified studies independently to determine whether an individual study was eligible for inclusion. The selection criteria for studies to be considered for this metaanalysis were as follows: 1) case–control or case–cohort study; 2) the RAD51135G>Cpolymorphism in cancer; 3) proper cancer diagnosis criteria; 4) original data; 5) not animal studies. The study would be excluded if the information could not be obtained.
Ethical consideration
The study has been approved by the Ethics committee of our Institutions.
Data extraction
The characteristics of selected studies were independently extracted through a standardized protocol by two authors, and the result was reviewed by a third investigator. The following information was sought from each study: first author, year of publication, study population (country, ethnicity), cancer types, the number of patients and controls for a study, genotype frequency for cases and controls, allele frequency in controls, and Hardy– Weinberg equilibrium (HWE).
Statistical analysis
Allele frequencies (–135C) at the RAD51 polymorphism from each respective study were determined by the allele counting method. Genotype distributions of controls were used to estimate the frequencies of the putative risk allele (–135C) using the inverse variance method [17,18]. The deviation from the Hardy–Weinberg Equilibrium (HWE) for distribution of the allele frequencies was analyzed by Fisher’s exact test in control groups, P < 0.05 was considered as representative of statistically significant. We examined the contrast of the C allele vs. G allele, CC vs. GG, CC vs. CG, and also examined the recessive genetic model (CC vs. CG+GG) and the dominant genetic model (CC+CG vs. GG). The associations between RAD51 (G135C) polymorphisms and cancer susceptibility were estimated by odds ratios (ORs) with 95% confidence intervals (CIs). The significance of the pooled OR was determined by the Z–test; P < 0.05 was considered statistically significant. Furthermore, to evaluate the ethnicity and cancer type–specific effects, subgroup analyses were performed.
Heterogeneity assumption was checked by a Chi–square based Q test, and it was considered statistically significant when P <0.1 [19]. Heterogeneity was also quantified with I2 metric (I2 = (Q–df)⁄Q×100%; I2 < 25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2=50–75%, large heterogeneity, I2>75%, extreme heterogeneity). When the effects were assumed to be homogenous (P > 0.1, I2 < 50%), the fixed–effects model was used; otherwise, the random–effects model was more appropriate. Sensitivity analysis was performed to evaluate the stability of the results. If more than seven studies were included, Begg’s test was used to measure the publication bias which was shown as a funnel plot [20]. P < 0.05 was considered as representative of statistically significant publication bias. All analyses were performed using the software STATA software, version 12.0 (Stata Corporation, College Station, TX,USA) and R statistical software, version 2.15.2 (http:⁄⁄www.r–project.org).
Results
Characteristics of studies
A total of fifty studies that met the inclusion concerning the association between RAD51135G>C polymorphism and risk of cancer were considered in the meta–analysis [12,15,16,21–67]. These studies involved 27,895 patients and 26,344 controls, containing thirty–eight Caucasian, five Asian, and seven mixed studies. In subgroup analysis, thirty–eight Caucasian studies (14,180⁄12,726) and five Asian studies (1,946⁄2,945) were included in ethnic–specific group. Additionally, twenty–six (19,716⁄19,735) studies focusing on breast cancer, seven (2,169⁄3,629) studies focusing on hematologic malignances, four (753⁄720) studies focusing on colorectal cancer, three (500⁄506) studies focusing on endometrial cancer, three (1,085⁄1,160) studies focusing on head and neck cancer, and two (2,925⁄1,749) studies focusing on ovarian cancer were also respectively evaluated. 84% (42⁄50) of these studies included used polymerase chain reactionrestriction fragment length polymorphism (PCR–RFLP) analysis for genotyping. Main characteristics of included studies were listed in Table 1.
Target
Drug
Major mechanism of action
Antidepressant effects
in human
Antidepressant effects
in rodents
BDNF levels
in human
BDNF levels
in rodents
Reference
NMDAR
Ketamine
Non-competitive antagonist
Yes
Yes
↑
↑
[67-72]
NMDAR
MK-0657
Selective NR2B antagonist
Yes
↑
[73]
NMDAR
Acamprosate
NMDA and mGluR5 antagonist
Inconclusive
Yes
↑
[73-75]
NMDAR
Memantine
Non-competitive low-affinity antagonist
No
↑
[76-78]
AMPAR
Ampakines
Positive allosteric modulator
Yes
Yes
↑
[79-85]
mGluR2/3
LY379268
Agonist
Clinical trials undergoing
Yes ↑ (enhancement with antidepressants)
↑ (with antidepressants)
[86-90]
mGluR2/3
LY341495
Antagonist
Clinical trials undergoing
Yes
↑ (enhancement with DOI)
[91-92]
mGluR5
MPEP
Selective antagonist
Yes
↑ (hippocampus) ↓ (cortex)
[93-96]
Other
Riluzole
Reduces extra-synaptic glutamate by inhibiting presynaptic release and enhancing glial uptake
Yes (preliminary)
↑
[97-99]
Table 1: Characteristics of the studies included in the meta-analysis.
Frequency of the C allele in different groups
The pooled RAD51–135C frequencies were 17.77 % (95 % CI: 17.29 – 18.25 %), and 32.49 % (95 % CI: 30.66 % – 34.32 %) in the controls of Caucasian, and Asian population. Genotype distributions in the controls of all studies were in agreement with HWE, except ten studies [10, 23–25, 27, 39, 48, 49, 58].
Results of meta–analysis
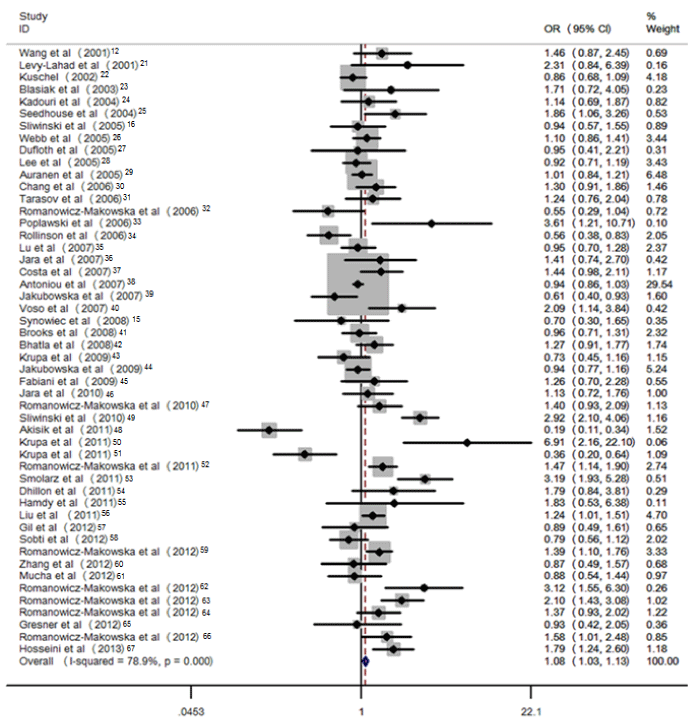
For each study, we investigated the association between the 135G>C polymorphism and risk of cancer. Overall, RAD51 135 C allele was associated with a statistically increased risk of cancer, compared with the G allele (OR =1.36 95% CI: 1.31–1.41) under random–effect model. Significant associations were also observed in the genetic models for CC vs. GG (OR =2.37 95% CI: 2.12–2.65), CC vs. CG (OR =4.02 95% CI: 3.62–4.46), recessive model (OR =3.74, 95% CI: 3.40–4.11), and dominant model (OR =1.08, 95% CI: 1.03– 1.13, Figure 1.). Z–test indicated that the pooled ORs were statistically significant.
Figure 1: Pooled OR (dominant model) and 95% CI of individual studies and pooled data for the association between polymorphism of RAD51 135G>C and cancer risk in overall population.
For each study, we investigated the association between the 135G>C polymorphism and risk of cancer. Overall, RAD51 135 C allele was associated with a statistically increased risk of cancer, compared with the G allele (OR =1.36 95% CI: 1.31–1.41) under random–effect model. Significant associations were also observed in the genetic models for CC vs. GG (OR =2.37 95% CI: 2.12–2.65), CC vs. CG (OR =4.02 95% CI: 3.62–4.46), recessive model (OR =3.74, 95% CI: 3.40–4.11), and dominant model (OR =1.08, 95% CI: 1.03– 1.13, Figure 1.). Z–test indicated that the pooled ORs were statistically significant.
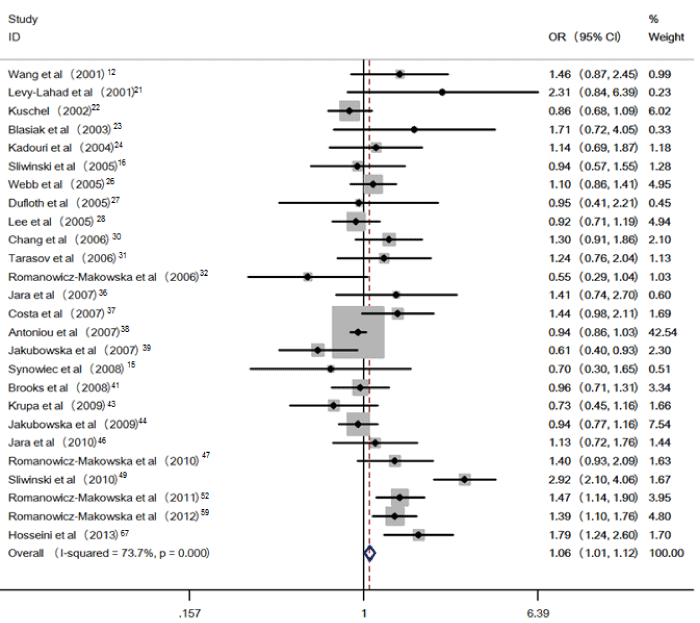
Additionally, the significant associations were found in the cancer subtypes including the breast cancer (C vs. G OR =1.32 95% CI: 1.26– 1.39; CC vs. GG OR=2.36, 95% CI 2.05–2.71; CC vs. CG OR=4.09, 95% CI 3.58–4.67; recessive model OR=3.73, 95% CI 3.31–4.21; and dominant model OR=1.06, 95% CI 1.01–1.12, Figure 3.), hematologic malignances (C vs. G OR =1.16 95% CI: 1.02–1.31; dominant model OR=1.18, 95% CI 1.03–1.36 ), colorectal cancer (C vs. G OR =1.62 95% CI: 1.37–1.91; CC vs. GG OR=2.06, 95% CI 1.48–2.87; CC vs. CG OR=3.74, 95% CI 2.72–5.15; recessive model OR=3.21, 95% CI 2.43–4.25 ), endometrial cancer (C vs. G OR =4.96 95% CI: 4.07–6.05; CC vs. GG OR=8.50, 95% CI 5.86–12.34; CC vs. CG OR=20.24, 95% CI 13.98–29.30; recessive model OR=13.96, 95% CI 10.25–19.02, and dominant model OR=2.39, 95% CI 1.74–3.29 ), and ovarian cancer (C vs. G OR =1.33 95% CI: 1.14–1.56; CC vs. GG OR=3.23, 95% CI 1.84–5.66; CC vs. CG OR=5.21, 95% CI 3.09–8.80; recessive model OR=5.50, 95% CI 3.37–8.98 ). The detailed results of meta–analysis were shown in Table 2.
Target
Drug
Major mechanism of action
Antidepressant effects
in human
Antidepressant effects
in rodents
BDNF levels
in human
BDNF levels
in rodents
Reference
NMDAR
Ketamine
Non-competitive antagonist
Yes
Yes
↑
↑
[67-72]
NMDAR
MK-0657
Selective NR2B antagonist
Yes
↑
[73]
NMDAR
Acamprosate
NMDA and mGluR5 antagonist
Inconclusive
Yes
↑
[73-75]
NMDAR
Memantine
Non-competitive low-affinity antagonist
No
↑
[76-78]
AMPAR
Ampakines
Positive allosteric modulator
Yes
Yes
↑
[79-85]
mGluR2/3
LY379268
Agonist
Clinical trials undergoing
Yes ↑ (enhancement with antidepressants)
↑ (with antidepressants)
[86-90]
mGluR2/3
LY341495
Antagonist
Clinical trials undergoing
Yes
↑ (enhancement with DOI)
[91-92]
mGluR5
MPEP
Selective antagonist
Yes
↑ (hippocampus) ↓ (cortex)
[93-96]
Other
Riluzole
Reduces extra-synaptic glutamate by inhibiting presynaptic release and enhancing glial uptake
Yes (preliminary)
↑
[97-99]
Table 2: Summary odds ratios (ORs) of the RAD51 135G/C polymorphism and cancer risk.
Sensitivity analysis
We conducted sensitivity analysis to evaluate the stability of the crude results which pooled with random–effects model. When any single study was deleted, the corresponding pooled ORs were not substantially altered (data not shown), suggesting that the results of this meta–analysis are stable.
Publication bias
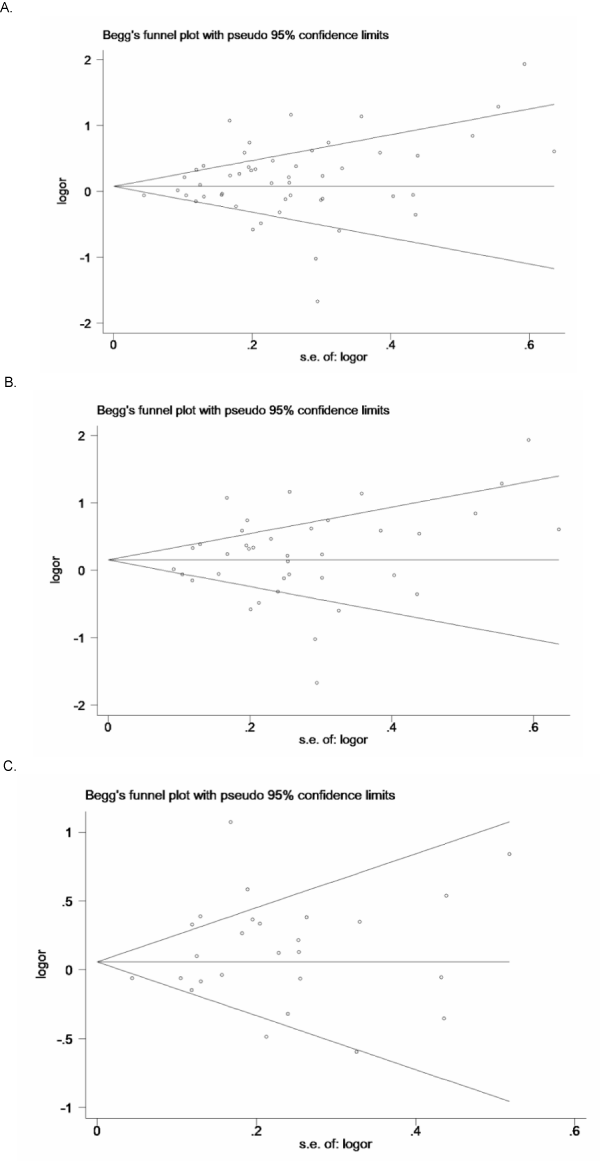
Begg’s test and a funnel plot were performed to assess the publication bias of the literature. The results indicated that no evidence of publication bias was detected in all the genetic models except for the recessive model in the breast cancer subgroup (Table 2, Figure 4A–C.).
Discussion
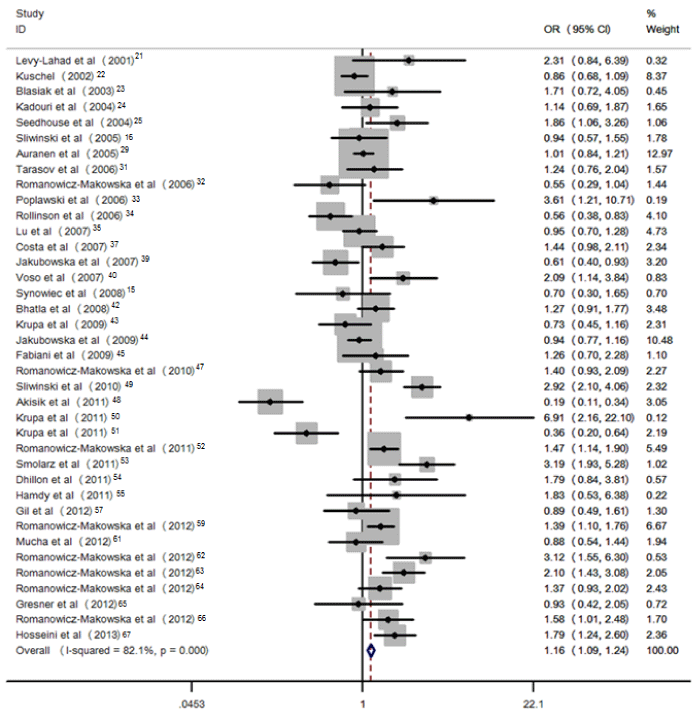
In the present study, we explored the association between the RAD51 135G>C polymorphism and cancer risk, involving fifty eligible case–control studies. In this meta–analysis, we collected a larger sample volume and examined the contrast of the C vs. G, CC vs. GG, CC vs. CG and also examined the recessive genetic model and the dominant genetic model. Furthermore, to evaluate the ethnicity and the disease based subtype–specific effects, subgroup analyses were performed. Our results indicated that the prevalenceof the C allele varied from 17.77 % to 32.49 % in different ethnic groups and individuals with the C allele have an increased risk of cancer in Caucasian population, but not in Asian population. In stratified analysis by cancer types, the significantly elevated risks with CC genotype were also found among breast cancer, hematologic malignances, colorectal cancer, endometrial cancer, and ovarian cancer.
Figure 2: Pooled OR (dominant model) and 95% CI of individual studies and pooled data for the association between polymorphism of RAD51 135G>C and cancer risk in Caucasian population.
Figure 3: Pooled OR (dominant model) and 95% CI of individual studies and pooled data for the association between polymorphism of RAD51 135G>C and breast cancer risk in cancer subgroup analysis.
Figure 4: Funnel plot of the RAD51 135G>C polymorphism and cancer risk in (A) overall population in dominant model (z = 1.26, P = 0.207), (B) Caucasian population in dominant model (z = 0.79, P = 0.428), (C) breast cancer (z = 0.37, P = 0.708) in dominant model.
RAD51 is a homologue of Escherichia coli recA protein, which is responsible for the central activity of the HR repair pathway. It catalyzes the invasion of the broken ends of the DSBs into the intact sister chromatid [68,69] The RAD51 gene containing 10 exons has been mapped to chromosome 15q15.1 [70]. The G>C polymorphism of 135–loci in RAD51 gene locating in the 5’UTR could affect mRNA stability, translation efficiency, protein level and finally influence the risk of cancer [71].
To date, a number of studies were performed to detect the association between RAD51 135G>C polymorphism and cancer risk. In order to evaluate the association in a larger population, some meta–analyses were performed to evaluate the association [72–75]. However, these previous meta–analyses have limitations in relatively small sample sizes and⁄or limited cancer type–specific analysis using the limited genetic models. Therefore, it is essential for us to perform a new updated meta–analysis to evaluate this association. Comparing with them, our study has some improvements. First, we enlarged the sample–size including all the cancer types. Second, we performed a more comprehensive data analysis including four different genetic models. Third, we made the subgroup analysis of ethnicity, cancer types. This is the first time to evaluate the relationships between RAD51 135G>C polymorphism and so many cancer types. Previous meta–analyses were carried out to assess the effect of RAD51 135G>C polymorphism on either the risk of breast cancer, or several limited cancer types only.
Though the results of this meta–analysis were powerful, some limitations still exist. First, it is clear that environmental factors play an important role in the etiology of cancer. However, the percentage of cancer caused by environmental factors is difficult to determine. The existence of gene–environment and gene–gene interactions may affect the accuracy of our results. Second, in the subgroup analyses, the involving number of population in Asians and other cancer types except for breast cancer were relatively small which may affect to explore the real associations. Third, this meta–analysis only focused on papers published in the English language and those which werereported in other languages might bias the present results. Fourth, the significance of heterogeneity among studies was observed. We pooled ORs with random–effects model in this condition. Sensitivity analysis suggested that the results of this meta–analysis are stable. Fifth, in our study, the studies including the number of GC+CC and GG only were also included, while they were deleted in some other meta–analysis. Finally, in our meta–analyses, we found the distribution of genotypes among controls was not agreement with HWE in some studies, which were included in this study. This may be due to chance, because studies with small sample size and selection bias may also contribute to the disaccord of HWE which may influence the risk effects. Other factors like differences in gene–gene and geneenvironment interactions from different genetic backgrounds and different matching criteria may also play a role in the discrepancy. In spite of these, when studies not in HWE were corrected to account for departures from HWE, then the pattern of results remained the same. And the result was also consistent with the most recently published meta–analysis [75], which excluded the studies in which genotypefrequencies in controls were not in accordance with HWE. Besides, our publication bias tests indicated there was no publication bias in RAD51 135G>C polymorphism, and it is likely to be reliable.
In conclusion, our result revealed that the C allele in 135–loci of RAD51 gene was associated with a significantly increased risk of cancers including breast cancer, hematologic malignances, colorectal cancer, endometrial cancer and ovarian cancer. The increased cancer risk was detected among Caucasian population, but not among Asian population. The effect of the variants on the expression levels and the possible functional role of the variants in cancer should be addressed in further studies.
Conflicts of Interest: We declare that there are no competing interests regarding the contents of this article.
Contributions
C.X. designed the study. C.X and B.B.Z. performed the literature search, data collection and data analysis. K.F.D, G.G. and L.M.L performed data gathering and quality assessment. All authors wrote and approved the manuscript.
References
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England journal of medicine 2000; 343: 78-85.
- Brody JG, Moysich KB, Humblet O, Attield KR, Beehler GP. Environmental pollutants and breast cancer: epidemiologic studies. Cancer. 2007; 109: 2667-2711.
- Hsu L, Zhao LP. Assessing familial aggregation of age at onset, by using estimating equations, with application to breast cancer. Am J Hum Genet. 1996; 58: 1057-1071.
- Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000; 26: 411-414.
- Hamilton AS, Mack TM. Puberty and genetic susceptibility to breast cancer in a case-control study in twins. N Engl J Med. 2003; 348: 2313-2322.
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008; 17: R109-115.
- Chung CC, Magalhaes WC, Gonzalez-Bosquet J, Chanock SJ. Genome-wide association studies in cancer--current and future directions. Carcinogenesis. 2010; 31: 111-120.
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M. A genome-wide association study identiies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007; 39: 870-874.
- Mohindra A, Hays LE, Phillips EN, Preston BD, Helleday T. Defects in homologous recombination repair in mismatch-repair-deicient tumour cell lines. Hum Mol Genet. 2002; 11: 2189-2200.
- Henning W, Stürzbecher HW. Homologous recombination and cell cycle checkpoints: Rad51 in tumour progression and therapy resistance. Toxicology. 2003; 193: 91-109.
- Dudás A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004; 566: 131-167.
- Wang WW, Spurdle AB, Kolachana P, Bove B, Modan B, et al. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and risk of cancer among BRCA1/2 mutation carriers. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2001; 10: 955- 960.
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31: 3406-3415.
- Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005; 219: 125-135.
- Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutat Res. 2008; 648: 65-72.
- Sliwinski T, Krupa R, Majsterek I, Rykala J, Kolacinska A. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat. 2005; 94: 105-109.
- Xuan C, Zhang BB, Yang T, Deng KF, Li M. Association between OCTN1/2 gene polymorphisms (1672C-T, 207G-C) and susceptibility of Crohn’s disease: a meta-analysis. Int J Colorectal Dis. 2012; 27: 11-19.
- Xuan C, Lun LM, Zhao JX, Wang HW, Zhu BZ. PTPN22 gene polymorphism (C1858T) is associated with susceptibility to type 1 diabetes: a meta-analysis of 19,495 cases and 25,341 controls. Ann Hum Genet. 2013; 77: 191-203.
- Xuan C, Zhang BB, Li M, Deng KF, Yang T. No association between APOE ε 4 allele and multiple sclerosis susceptibility: a meta-analysis from 5472 cases and 4727 controls. J Neurol Sci. 2011; 308: 110-116.
- Xuan C, Bai XY, Gao G, Yang Q, He GW. Association between polymorphism of methylenetetrahydrofolate reductase (MTHFR) C677T and risk of myocardial infarction: a meta-analysis for 8,140 cases and 10,522 controls. Arch Med Res. 2011; 42: 677-685.
- Levy-Lahad E, Lahad A, Eisenberg S, Dagan E, Paperna T. A single nucleotide polymorphism in the RAD51 gene modiies cancer risk in BRCA2 but not BRCA1 carriers. Proc Natl Acad Sci U S A. 2001; 98: 3232-3236.
- Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002; 11: 1399-1407.
- Blasiak J, Przybyłowska K, Czechowska A, Zadrozny M, Pertyński T. Analysis of the G/C polymorphism in the 5’-untranslated region of the RAD51 gene in breast cancer. Acta Biochim Pol. 2003; 50: 249-253.
- Kadouri L, Kote-Jarai Z, Hubert A, Durocher F, Abeliovich D. A single- nucleotide polymorphism in the RAD51 gene modiies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br J Cancer. 2004; 90: 2002-2005.
- Seedhouse C, Faulkner R, Ashraf N, Das-Gupta E, Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clinical cancer research : an oficial journal of the American Association for Cancer Research 2004; 10: 2675-2680.
- Webb PM, Hopper JL, Newman B, Xiaoqing Chen, Livia Kelemen, et al. Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2005; 14: 319-323.
- Duloth RM, Costa S, Schmitt F, Zeferino LC. DNA repair gene polymorphisms and susceptibility to familial breast cancer in a group of patients from Campinas, Brazil. Genet Mol Res. 2005; 4: 771-782.
- Eckhardt BL, Parker BS, van Laar RK, Restall CM, Natoli AL, et al. Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Molecular cancer research : MCR 2005; 3: 1-13.
- Auranen A, Song H, Waterfall C, Dicioccio RA, Kuschel B. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005; 117: 611-618.
- Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006; 15: 754-761.
- Tarasov VA, Aslanyan MM, Tsyrendorzhiyeva ES, Litvinov SS, Gar’kavtseva RF, et al. Genetically determined subdivision of human populations with respect to the risk of breast cancer in women. Doklady biological sciences: proceedings of the Academy of Sciences of the USSR, Biological sciences sections / translated from Russian 2006; 406: 66-69.
- Romanowicz-Makowska H, Smolarz B, Zadrozny M, Kulig A. Analysis of RAD51 polymorphism and BRCA1 mutations in Polish women with breast cancer. Exp Oncol. 2006; 28: 156-159.
- Poplawski T, Arabski M, Kozirowska D, Blasinska-Morawiec M, Morawiec Z. DNA damage and repair in gastric cancer--a correlation with the hOGG1 and RAD51 genes polymorphisms. Mutat Res. 2006; 601: 83-91.
- Rollinson S, Smith AG, Allan JM, Adamson PJ, Scott K, et al. RAD51 homologous recombination repair gene haplotypes and risk of acute myeloid leukaemia. Leukemia research 2007; 31: 169-174.
- Lu J, Wang LE, Xiong P, Sturgis EM, Spitz MR. 172G>T variant in the 5’ untranslated region of DNA repair gene RAD51 reduces risk of squamous cell carcinoma of the head and neck and interacts with a P53 codon 72 variant. Carcinogenesis. 2007; 28: 988-994.
- Jara L, Acevedo ML, Blanco R, Castro VG, Bravo T. RAD51 135G>C polymorphism and risk of familial breast cancer in a South American population. Cancer Genet Cytogenet. 2007; 178: 65-69.
- Costa S, Pinto D, Pereira D, et al. DNA repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: a study on a Portuguese population. Breast cancer research and treatment 2007; 103: 209-217.
- Antoniou AC, Sinilnikova OM, Simard J, Léoné M, Dumont M. RAD51 135G-->C modiies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007; 81: 1186-1200.
- Jakubowska A, Gronwald J, Menkiszak J, Górski B, Huzarski T, et al. The RAD51 135 G>C polymorphism modiies breast cancer and ovarian cancer risk in Polish BRCA1 mutation carriers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2007; 16: 270- 275.
- Voso MT, Fabiani E, D’Alo’ F, Guidi F, Di Ruscio A. Increased risk of acute myeloid leukaemia due to polymorphisms in detoxiication and DNA repair enzymes. Ann Oncol. 2007; 18: 1523-1528.
- Brooks J, Shore RE, Zeleniuch-Jacquotte A, Currie D, Afanasyeva Y, et al. Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2008; 17: 1016-1019.
- Bhatla D, Gerbing RB, Alonzo TA, Mehta PA, Deal K. DNA repair polymorphisms and outcome of chemotherapy for acute myelogenous leukemia: a report from the Children’s Oncology Group. Leukemia. 2008; 22: 265-272.
- Krupa R, Synowiec E, Pawlowska E, Morawiec Z, Sobczuk A. Polymorphism of the homologous recombination repair genes RAD51 and XRCC3 in breast cancer. Exp Mol Pathol. 2009; 87: 32-35.
- Jakubowska A, Jaworska K, Cybulski C, Janicka A, Szymańska-Pasternak J. Do BRCA1 modiiers also affect the risk of breast cancer in non-carriers? Eur J Cancer. 2009; 45: 837-842.
- Fabiani E, D’Alò F, Scardocci A, Greco M, Di Ruscio A. Polymorphisms of detoxiication and DNA repair enzymes in myelodyplastic syndromes. Leuk Res. 2009; 33: 1068-1071.
- Jara L, Dubois K, Gaete D, de Mayo T, Ratkevicius N. Variants in DNA double-strand break repair genes and risk of familial breast cancer in a South American population. Breast Cancer Res Treat. 2010; 122: 813-822.
- Romanowicz H, Smolarz B, Baszczynski J, Zadrozny M, Kulig A. Genetics polymorphism in DNA repair genes by base excision repair pathway (XRCC1) and homologous recombination (XRCC2 and RAD51) and the risk of breast carcinoma in the Polish population. Polish journal of pathology : oficial journal of the Polish Society of Pathologists 2010; 61: 206-212.
- Akisik E, Yazici H, Dalay N. ARLTS1, MDM2 and RAD51 gene variations are associated with familial breast cancer. Mol Biol Rep. 2011; 38: 343-348.
- Sliwinski T, Sitarek P, Stetkiewicz T, Sobczuk A, Blasiak J. Polymorphism of the ERalpha and CYP1B1 genes in endometrial cancer in a Polish subpopulation. J Obstet Gynaecol Res. 2010; 36: 311-317.
- Krupa R, Sobczuk A, Popławski T, Wozniak K, Blasiak J. DNA damage and repair in endometrial cancer in correlation with the hOGG1 and RAD51 genes polymorphism. Mol Biol Rep. 2011; 38: 1163-1170.
- Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, Wasylecka M. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer--a case control study. Mol Biol Rep. 2011; 38: 2849-2854.
- Romanowicz-Makowska H, Smolarz B, Zadrozny M, Boguslaw Westfal, Jakub Baszczynski, et al. Single nucleotide polymorphisms in the homologous recombination repair genes and breast cancer risk in Polish women. The Tohoku journal of experimental medicine 2011; 224: 201-208.
- Smolarz B, Samulak D, Michalska M, Góralczyk B, Szyłło K. 135G>C and 172G>T polymorphism in the 5’ untranslated region of RAD51 and sporadic endometrial cancer risk in Polish women. Pol J Pathol. 2011; 62: 157-162.
- Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia--results of a pilot study. Urol Oncol. 2011; 29: 641-646.
- Hamdy MS, El-Haddad AM, Bahaa El-Din NM, Makhlouf MM, Abdel-Hamid SM. RAD51 and XRCC3 gene polymorphisms and the risk of developing acute myeloid leukemia. Journal of investigative medicine : the oficial publication of the American Federation for Clinical Research 2011; 59: 1124-1130.
- Liu L, Yang L, Mi Y, Wang J, Li J. RAD51 and XRCC3 polymorphisms: impact on the risk and treatment outcomes of de novo inv(16) or t(16;16)/CBFβ- MYH11(+) acute myeloid leukemia. Leuk Res. 2011; 35: 1020-1026.
- Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012; 39: 527-534.
- Sobti RC, Kaur S, Sharma VL, Singh SK, Hosseini SA. Susceptibility of XPD and RAD51 genetic variants to carcinoma of urinary bladder in North Indian population. DNA Cell Biol. 2012; 31: 199-210.
- Romanowicz-Makowska H, Smolarz B, Zadrozny M, B Westfa, J Baszczynski, et al. The association between polymorphisms of the RAD51-G135C, XRCC2- Arg188His and XRCC3-Thr241Met genes and clinico-pathologic features in breast cancer in Poland. European journal of gynaecological oncology 2012; 33: 145-150.
- Zhang L, Ruan Z, Hong Q, Gong X, Hu Z. Single nucleotide polymorphisms in DNA repair genes and risk of cervical cancer: A case-control study. Oncol Lett. 2012; 3: 351-362.
- Mucha B, Przybyłowska-Sygut K, Dziki L, Dziki A, Sygut A. Lack of association between the 135G/C RAD51 gene polymorphism and the risk of colorectal cancer among Polish population. Pol Przegl Chir. 2012; 84: 358- 362.
- Romanowicz-Makowska H, Smolarz B, Samulak D, Michalska M, Lewy J. A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and ovarian cancer risk in Polish women. Eur J Gynaecol Oncol. 2012; 33: 406- 410.
- Romanowicz-Makowska H, Samulak D, Michalska M, Sporny S, Langner E. RAD51 gene polymorphisms and sporadic colorectal cancer risk in Poland. Pol J Pathol. 2012; 63: 193-198.
- Romanowicz-Makowska H, Smolarz B, Gajęcka M, Kiwerska K, Rydzanicz M. Polymorphism of the DNA repair genes RAD51 and XRCC2 in smoking- and drinking-related laryngeal cancer in a Polish population. Arch Med Sci. 2012; 8: 1065-1075.
- Gresner P, Gromadzinska J, Polanska K, Twardowska E, Jurewicz J. Genetic variability of Xrcc3 and Rad51 modulates the risk of head and neck cancer. Gene. 2012; 504: 166-174.
- Romanowicz-Makowska H, Smolarz B, Polac I, Sporny S. Single nucleotide polymorphisms of RAD51 G135C, XRCC2 Arg188His and XRCC3 Thr241Met homologous recombination repair genes and the risk of sporadic endometrial cancer in Polish women. The journal of obstetrics and gynaecology research 2012; 38: 918-924.
- Hosseini M, Houshmand M, Ebrahimi A . RAD51 polymorphisms and breast cancer risk. Mol Biol Rep. 2013; 40: 665-668.
- Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001; 477: 131-153.
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006; 25: 222-231.
- Hasselbach L, Haase S, Fischer D, Kolberg HC, Stürzbecher HW. Characterisation of the promoter region of the human DNA-repair gene Rad51. Eur J Gynaecol Oncol. 2005; 26: 589-598.
- Gray NK. Translational control by repressor proteins binding to the 5’UTR of mRNAs. Methods Mol Biol. 1998; 77: 379-397.
- Zhou GW, Hu J, Peng XD, Li Q. RAD51 135G>C polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2011; 125: 529-535.
- Yu KD, Yang C, Fan L, Chen AX, Shao ZM. RAD51 135G>C does not modify breast cancer risk in non-BRCA1/2 mutation carriers: evidence from a meta- analysis of 12 studies. Breast Cancer Res Treat. 2011; 126: 365-371.
- Wang Z, Dong H, Fu Y, Ding H. RAD51 135G>C polymorphism contributes to breast cancer susceptibility: a meta-analysis involving 26,444 subjects. Breast Cancer Res Treat. 2010; 124: 765-769.
- Zhao M, Chen P, Dong Y, Zhu X, Zhang X. Relationship between Rad51 G135C and G172T Variants and the Susceptibility to Cancer: A Meta- Analysis Involving 54 Case-Control Studies. PLoS One. 2014; 9: e87259.