Research Article
Austin J Pharmacol Ther. 2014; 2 (3). 1020
Salinomycin Suppresses PDGFRβ, MYC, and Notch Signaling in Human Medulloblastoma
Shuang Zhou1, Fengfei Wang1, Ying Zhang1, Max R Johnson2, Steven Qian1, Min Wu3, Erxi Wu1*
1Department of Pharmaceutical Sciences, North Dakota State University, USA
2Retina Consultants Ltd and University of North Dakota, USA
3Department of Basic Sciences, University of North Dakota, USA
*Corresponding author: : Erxi Wu, Department of Pharmaceutical Sciences, North Dakota State University, Fargo, ND, USA
Received: February 15, 2014; Accepted: April 20, 2014; Published: April 22, 2014
Abstract
Medulloblastoma (MB) is the most common childhood brain tumor. Despite improved therapy and management, approximately 30% of patients die of the disease. To search for a more effective therapeutic strategy, the effects of salinomycin were tested on cell proliferation, cell death, and cell cycle progression in human MB cell lines. The results demonstrated that salinomycin inhibits cell proliferation, induces cell death , and disrupts cell cycle progression in MB cells. Salinomycin was also tested on the expression levels of key genes involved in proliferation and survival signaling and revealed that salinomycin down–regulates the expression of PDGFRβ, MYC, p21 and Bcl–2 and up–regulates the expression of cyclin A. In addition, the results reveal that salinomycin suppresses the expression of Hes1 and Hes5 in MB cells. Our datashed light on the potential of using salinomycin as a novel therapeutic agent for patients with MB.
Key words: Salinomycin; Medulloblastoma; PDGFRβ; MYC; Notch Signaling
Abbreviations
PDGFRβ: Beta–type Platelet–Derived Growth Factor Receptor; Bcl–2: B–cell Lymphoma 2; DLL1: Delta–Like 1 (Drosophila); Dll3: Delta–Like 3 (Drosophila); Hes1: Hairy And Enhancer Of Split 1 (Drosophila); Hes5: Hairy And Enhancer of Split 5 (Drosophila); Hey1: Hairy⁄Enhancer–of–Split Related with Yrpw Motif 1; Hey2: Hairy⁄Enhancer–of–Split Related with Yrpw Motif 2; Dtx1: Deltex Homolog 1 (Drosophila); Dtx2: Deltex Homolog 2 (Drosophila); MAML1: Mastermind–Like 1 (Drosophila); MAML2: Mastermind– Like 2 (Drosophila); MAML3: Mastermind–Like 3 (Drosophila); RBPJ: Recombination Signal Binding Protein for Immunoglobulin Kappa J Region; MTS: 3–(4, 5–Dimethylthiazol–2–Yl)–5–(3– Carboxymethoxyphenyl)–2–(4–Sulfophenyl)–2H– Tetrazolium, Inner Salt; DMSO: Dimethyl Sulfoxide.
Introduction
Medulloblastoma (MB), an embryonal neuroepithelial tumor of the cerebellum, is the most common malignant brain tumor in children [1]. This highly invasive tumor has a tendency to disseminate throughout the central nervous system early in its course. Although, the medical treatment outcome for children with MB has improved over the past several decades, approximately one–third of patients with MB tumors remain incurable. Moreover, current medical treatments have associated toxicities that can cause significant disabilities in long–term survivors [2].Thus, more effective drugs are needed for treating patients with MB.
Salinomycin is a 751 Da mono carboxylic polyether antibiotic which is widely used as an anti–coccidial drug. Recently, salinomycin has been found to reduce the proportion of breast cancer stem cells (CSC) by more than 100–fold compared to paclitaxel, a common drug used for breast cancer [3]. Cumulative findings strongly suggest that salinomycin is a selective killer of human CSC and an effective killer of multi–drug resistant human CSC–like cells [4–11]. The action mechanism of salinomycin in cancer and CSCs has been shown to modulate multiple signaling pathways including the Wnt, NF–κB, and p38 MAPK pathways [12–14].
MB cells have the potential to differentiate to neuronal and⁄or glial cells [15,16], indicating that MB cells are of stem cell origin. Growing evidence points toward the existence of a CSC–like population that may contribute to MB therapy resistance [17–19]. Notch signaling is critical for cell differentiation and proliferation and plays a fundamental role in MB initiation and progression by regulating downstream effectors, e.g., MYC [20–22]. The Notch pathway inhibitors, e.g., γ–secretase inhibitor MK–0752, suppress the cleavageof Notch, eliminate the stem cell like population [23–25], reduce cell proliferation and increase apoptosis [23–25], which implicates Notch signaling as a target that may constitute an additional promising treatment strategy for MB patients. In the present study, for the first time, we determined the anticancer effects of salinomycin in 3 MB cell lines. We also measured the effects of salinomycin on the expression of a few genes critical in cell proliferation, survival, and differentiation in MB cells.
Materials and methods
Cell Culture and Chemicals
MB cell lines (Daoy and D283) were obtained from ATCC and D425 cells were a gift from Dr. Darell D. Bigner [26]. Daoy and D283 MB cells were maintained in minimum Essential Medium (MEM) (Cellgro) supplemented with 4 mM L–glutamine, 100 units⁄ ml penicillin, 100 µg⁄ml streptomycin, 1% sodium pyruvate, 1% nonessential amino acids, and 10% fetal bovine serum (FBS) at 37°C with 5% CO2. D425 cells were maintained in improved MEM (Zinc Optioin 1 x) (GIBCO) supplemented with 10% FBS at 37°C with 5% CO2. Salinomycin and propidium iodide (PI) were obtained from Sigma.
Cell Proliferation Analysis
MB cells (Daoy 1x104⁄well, D283 5 x104⁄well, D425 1 x105⁄well) were placed in 96–well plates overnight. Salinomycin solution or identical volume of control (DMSO) was added to the appropriate wells. After 48 hours of treatment, a 20–µl of MTS solution (Promega) was added to each well. The cell number in each condition was determined by measuring the optical densities at 490 nm after 4–hour of incubation. The results were expressed as the percentages of control cultures.
Cell Death and Cell Cycle Analysis
MB cells (5 x105 ⁄well) were placed in 6–well plates overnight. Salinomycin or identical volume of control was added to the appropriate wells. After 24 hours of treatment, the cells were stained with PI and analyzed for cell death and cell cycle distribution using the Cell Lab Quanta TM SC system (Beckman Coulter) followed by Flow Cytometry Analysis (FACS).
Western Blotting Analysis
MB cells were harvested at the 24–hour time point for protein by adding Tris–triton cell lysis buffer (1% Triton, 50 mM Tris–HCl pH 7.4, 10% glycerol, 150 mM NaCl) supplemented with protease inhibitor cocktail (Roche Applied Science) and phosphates inhibitor cocktail 100x (cell signaling). The protein samples were separated using a 10%–12% SDS–PAGE gel, and then transferred onto a nitrocellulose membrane. Immunoblots were probed with antibodies specific for cyclin A, Bcl–2, p–21 (Santa Cruz), PDGFRβ (Epitomic), and MYC (Cell signaling). β–actin (Sigma) served as a loading control. Signals of the specific proteins were detected by using the Immun– Star HRP peroxide Luminol⁄Enhancer kit (BIO–RAD) and recorded on KODAK Biomax light film.
RT–PCR Analysis
RNA was isolated from MB cells using the RNeasy Plus (Qiagen) by following the manufacturer’s protocol. The quantity and purity of RNA were determined using a Nano Drop 1000 spectrophotometer (Thermo Scientific). 1 µg of total RNA was used to prepare cDNA using Super Script first–strand synthesis system (Invitrogen) by following the instructions provided by the manufacturer. Primers used in this study are listed in Table 1 and synthesized by Integrated DNA Technology. Semi–quantitative PCR was achieved by amplifying genes using an equal amount of cDNA and limited number of cycles. For detection of basal levels, 35 cycles were used for all genes except GAPDH. For detection the effects of salinomycin on gene expression, PCR conditions are listed in Table 1. PCR amplification was performed using GoTaq Hot Start Colorless Master Mix (Promega) and MJ Mini Personal Thermal Cycler (Bio–Rad).The results were visualized by analyzing the samples using DNA gel.
Gene
Primer Sequences (5’->3’)
Sizes of PCR products (bp)
Annealing temperature (°C)
Cycle of amplification
NOTCH1
Forward: GACAGCCTCAACGGGTACAA
Reverse: CACACGTAGCCACTGGTCAT
137
55
40
NOTCH2
Forward: GGAGCTACTGTGAGGAGCAA
Reverse: GATTTCATACCCCGAGTGCC
238
55
35
NOTCH3
Forward: TGTCAACGAGTGTCTGTCGG
Reverse: TTGACTCGGTCCTTGCAGAC
174
55
35
NOTCH4
Forward: AGTGAGAGCTCTGAGGGTCC
Reverse: TGGGTCTGACCACTGAGACA
137
55
35
JAG1
Forward: GGCTGCAATAAGTTCTGCCG
Reverse: CAGCCTTGTCGGCAAATAGC
131
55
35
JAG2
Forward: TGCAAAAACCTGATTGGCGG
Reverse: CACACACTGGTACCCGTTCA
144
60
35
DLL1
Forward: ACCTCGCAACAGAAAACCCA
Reverse: GTGTTCGTCACACACGAAGC
146
55
35
DLL3
Forward: CCGAGCTCGTCCGTAGATTG
Reverse: AGGGTAGGGAAAAAGCAGGTG
165
55
35
DLL4
Forward: TTAAGCACTTCCAGGCGGTC
Reverse: GATGAGCGAGAAGGTACCCG
170
55
35
HES1
Forward: AAGAAAGATAGCTCGCGGCA
Reverse: CCTCGGTATTAACGCCCTCG
208
55
40
HES5
Forward: GAGAAAAACCGACTGCGGAA
Reverse: TAGTCCTGGTGCAGGCTCTT
221
60
35
HEY1
Forward: TAATTGAGAAGCGCCGACGA
Reverse: GCTTAGCAGATCCTTGCTCCA
108
60
35
HEY2
Forward: AGATGCTTCAGGCAACAGGG
Reverse: GCGCAACTTCTGTTAGGCAC
102
55
35
DTX1
Forward: ACTTGAATGGTACTGGGCCG
Reverse: CACATCCTCGGGATTCTTACTCTT
221
55
35
DTX2
Forward: AGATTTGCCCGGTTTTTGTTG
Reverse: TCCGGCAGATCTTTTCTCTCTG
136
55
35
MAML1
Forward: CACGAGCAGAACTCCCTGTT
Reverse: CAGGGACACTGGAAGGGTTC
103
55
40
MAML2
Forward: ACAACCCTATGATGCCACGG
Reverse: CCCAGTTTGGTGCAGTTGTG
189
55
40
MAML3
Forward: ATAGGACCCTCCCAGAACCC
Reverse: CCCTGGGCTTGGTTATGTGT
185
55
35
RBPJ
Forward: CAGTGCTGGATCTGGGAATCT
Reverse: AATTTCCCAGGCGATGGAGC
188
61
35
NRARP
Forward: CACCAGGACATCGTGCTCTA
Reverse: GTAGTTGGCGGGAAGGTACA
128
55
35
GAPDH
Forward: GAGTCAACGGATTTGGTCGT
Reverse: TTGATTTTGGAGGGATCTCG
237
57
25
Table 1: Primers and conditions used for the semi-quantitative RT-PCR analysis.
Statistical Analysis
All quantitative data are represented as mean ± standard deviation (SD). Statistical tests were performed using the Minitab 16.1.1 software package. Comparisons between two groups were carried out using paired student’s t test and p < 0.05 was considered as statistical significance.
Results
Salinomycin suppresses cell proliferation, and induces cell death and S⁄G2 cell cycle arrest in MB cells
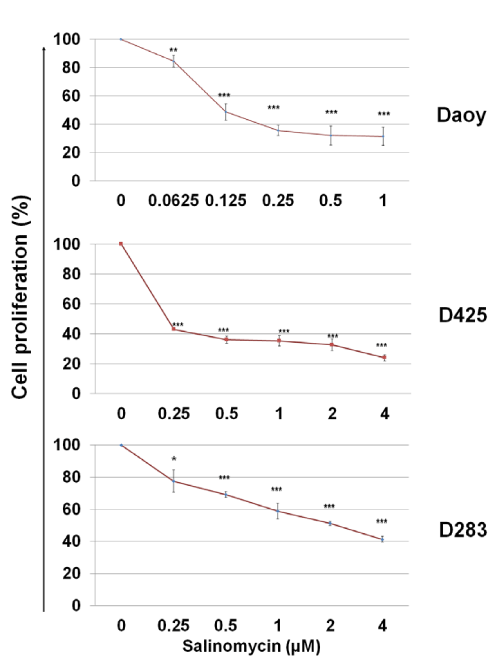
To assess the cytotoxicity of salinomycin on MB cells, Daoy, D425, and D283 cells were treated with salinomycin for 48 hours at indicated concentrations and then, the rates of cell proliferation were determined by using a MTS assay. As shown in Figure. 1, salinomycin suppressed MB cell proliferation in a dose–dependent manner. The IC50 of salinomycin are 0.1 µM, 0.25 µM, and 2 µM for Daoy, D425, and D283 cells, respectively (Figure. 1).
Figure 1: Effects of salinomycin on MB cell proliferation. Cells in complete medium were treated with salinomycin. After 48 hours, the number of viable cells in each well was determined using an MTS assay (Promega). The optical densities were measured at 490 nm. The results were calculated as the percentage against control cultures and presented as mean ± SD. The statistical differences were determined using paired student’s t–test and performed using Minitab. * p<0.05, **p<0.01, and ***p<0.001.
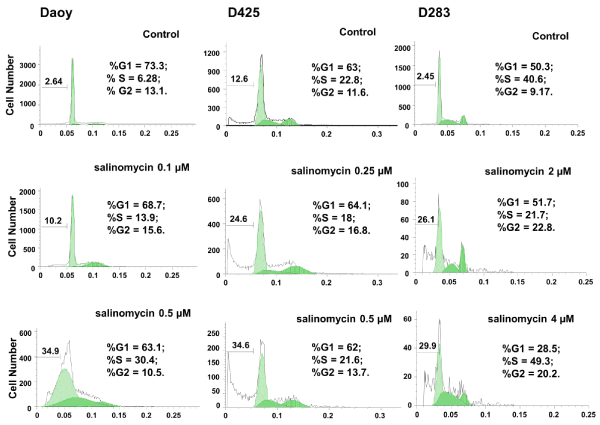
To further determine the effects of salinomycin on MB cells, Daoy, D425, and D283, cells were treated with salinomycin at the concentrations indicated in Figure. 2. After 24 hours of treatment, cells were analyzed for cell viability and cell cycle progression using PI staining followed by FACS analysis. As shown in Figure. 2, an increase in cell death (sub–G0) was observed in all three cell lines in response to salinomycin treatment. In addition, salinomycin induced Daoy cells arrest at S⁄G2 phases and D425 and D283 cells at G2 phase under low concentration. Under high concentration, with exception of D425 which showed little change, Daoy and D283 cells were both arrested at S phase (Figure. 2).
Figure 2: Salinomycin induces apoptosis and affects cell cycle progression in MB cell lines. Cells were treated with salinomycin for 24 hours and analyzed for cell cycle distribution and cell death. Cell cycle and cell death are analyzed independently using FlowJo. The cell death percentage is calculated against all cell population including apoptotic cells and live cells in cell cycle phases.
Salinomycin suppresses the expression of genes involved in MB proliferation and metastasis
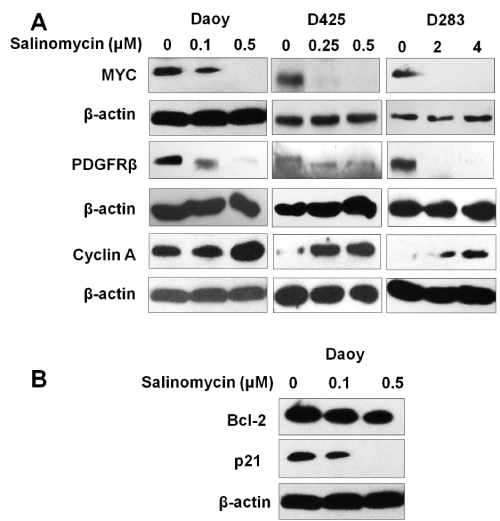
MYC and PDGFRβ were previously reported to be involved in MB growth and metastasis [27–30]. To understand the action mechanism of salinomycin on MB, the effects of salinomycin were examined on the expression of MYC and PDGFRβ in MB cells. Our results showed that all three MB cell lines, Daoy, D283, and D425, expressed high levels of PDGFRβ and MYC. Notably, after treatment with salinomycin at the concentration indicated in Figure. 3 for 24 hours, we observed a markedly down–regulation of both MYC and PDGFRβ in all 3 tested MB cell lines (Figure. 3A).
Figure 3: Effects of salinomycin on the expression levels of key regulators of cell proliferation, cell cycle progression and cell death. (A) Salinomycin down–regulates MYC and PDGFRβ, up–regulates cyclin A. (B) Salinomycin down–regulates the expression level of Bcl–2 and p21. Cells were treated with salinomycin for 24 hours at indicated concentrations and analyzed by Western blotting (β–actin was used as an internal loading control).
Salinomycin treatment has differential effects on the expression levels of cyclin A, p21, and Bcl–2
To further elucidate the mechanism of salinomycin’s action, the expression levels of key regulators of cell cycle progression and apoptosis were examined in response to salinomycin treatment by western blotting. The results showed that salinomycin treatment markedly increased cyclin a expression (Figure 3A). This effect could be due to the prolonged S⁄G2 phases. In addition, reduced antiapoptotic protein p21 and the survival protein Bcl–2 in MB cells were observed in response to salinomycin treatment (Figure. 3B). The modulation of cyclin A and p21 may contribute to a reduction in cell proliferation and the down regulation of Bcl–2 and p21 may contribute to salinomycin induced cell death.
Suppression of Notch signaling by salinomycin in MB cells
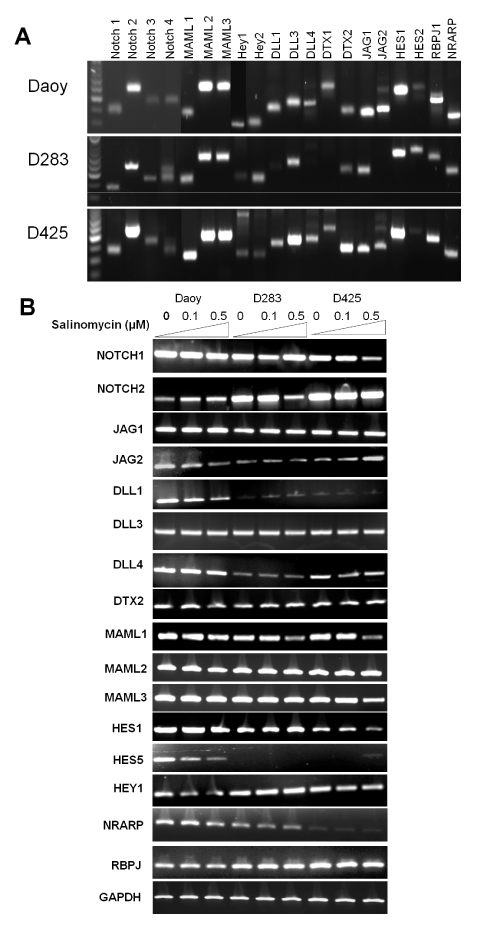
To evaluate the effects of salinomycin on the expression levels of genes in Notch signaling in MB cells, we first determined the basal levels of genes in Notch signaling using RT–PCR analysis. As shown in Figure. 4A, all 3 cell lines expressed relatively high levels of Notch 2, JAG1, MAML1–3, DLL3, Hes1, RBPJ1, NRARP. The effects of salinomycin on the transcription of key genes in Notch signaling such as Notch receptors (e.g., Notch 1 and 2), Notch ligands (e.g., JAG1 and DLL1), transcriptional co–activators for Notch signaling (e.g., MAML 1–3), and Notch signaling effectors (e.g., Hes1, Hes5, and HEY1) [31,32], were assessed. We observed an inhibition of Notch signaling by salinomycin in all tested cell lines that manifested suppression on transcription of DLL1, MAML1, Hey1, Hes1, and Hes5 genes (Figure. 4B).
Figure 4: Salinomycin modulates Notch signaling in MB cells. (A) The basal levels of genes in Notch signaling in the three MB cells lines. (B) Salinomycin down–regulated the mRNA levels of key components in Notch signaling in MB cells. Cells were treated with salinomycin for 24 hours at indicated concentrations and analyzed by RT–PCR analysis (β–actin was used as an internal loading control).
Discussion
In this study, we found that salinomycin at the concentration range of 0.25–4 µM significantly inhibits cell proliferation and induces cell death and cell cycle arrest. Through analyzing of changes in expression of genes and⁄or proteins that are involved in cell proliferation, cell death, and the Notch signaling pathway in response to salinomycin treatment, we reveal that salinomycin suppresses the expression of PDGFRβ, MYC, Bcl–2, p21 and some key effectors in the Notch signaling pathway (e.g., Hes1).
Since the discovery that salinomycin has anti–CSC activity in breast cancer [3], several recent studies have shown that salinomycin possesses profound anti–cancer and anti–CSC activities in other cancer types in vitro, and in vivo xenografted mouse models, as well as pilot clinical studies in patients [33–35]. Nevertheless, the effects of salinomycin on MB cells have not been previously studied. In the present study, we demonstrate that salinomycin hasprofound cytotoxicity against human MB cells. This conclusion was supported by a dose–dependent increase of cell death (the sub–G0 population) and a significant reduction of cell proliferation upon salinomycin treatment. Cyclin A is required for DNA replication in both S and G2 phases [36], the up–regulated cyclin A levels might be due to the prolonged S⁄G2 phases by salinomycin treatment. In addition, the cell cycle arrest at S⁄G2 phases and up–regulation of cyclin A expression were well correlated with cell proliferation data.
Hes1 and Hes 5 are critical effectors of the Notch signaling pathway which plays an important role in MB disease progression and patient survival. Fan et al. have reported that the Hes1 expression activated by Notch signaling is associated with significantly lower survival in MB patients [24]. Research from the same group also revealed that the blockade of the Notch pathway suppressed Hes1 expression and can cause cell apoptosis, cell cycle exit, and differentiation in MB cells [23]. This research suggested a role of Notch signaling in MB CSC maintenance. Our data show that salinomycin downregulated the transcription of both Hes1 and DLL1. This partially explained the effects of salinomycin on MB cell survial and indicatedits role on MB CSC maintenance. In addition, MAML1 which was previously shown as a coactivator to amplify the Notch induced transcription of Hes1 was also inhibited by salinomycin in MB cells [37]. The suppression of the gene expression in Notch signaling may also partially explain the downregulated protein levels of p21 which is also a target gene of Notch signaling [38].
It has been noticed that MYC is a downstream target of canonical Wnt signaling [39]. Indeed, salinomycin blocks the phosphorylation of the Wnt co–receptor lipoprotein receptor related protein 6 (LRP6) and induces its degradation in Wnt–transfected HEK293 cells [13]. It is possible that the down–regulated MYC might be partially caused by salinomycin’s impact on Wnt signaling. In this study, we have observed the suppression on Notch signaling by salinomycin in MB cells and MYC is also a target molecule of Notch signaling [40,41]. The possibility exists that salinomycin down–regulates MYC at least partially via Notch signaling. In addition, MYC is a downstream of PDGFR signaling [42,43]. Therefore, targeting these pathways simultaneously for MB should provide an effective strategy for the treatment of MB.
MYC is commonly deregulated in MB [44–46] and modulates multiple cellular events through alteration of the expression of a number of functionally important target genes [47]. Recent studies show that among the four subtypes of MB, the Group 3 MB chracterized with MYC overexpression indicates aggressive disease and poor prognosis [48]. Moreover, blocking MYC significantly reduces MB cell growth [49]. In addition to MYC, high levels of PDGFRβ have also been correlated with an aggressive phenotype of MB [50]. In this study, by showing that salinomycin could suppress the expression of MYC and PDGFRβ at the same time, our results uncover a new mechanistic aspect of salinomycin’s potent anti–cancer effects and highlight the value of salinomycin as a very promising drug for treating MB [51].
Conclusion
Our study demonstrates that salinomycin exhibits cytotoxic effects in human MB cells. Our data reveal that the treatment with salinomycin is effective in inhibiting MB cell proliferation and cell cycle progression and inducing cell death. We also show that MB cells have altered multiple signaling pathways after salinomycin treatment. The down–regulation of PDGFRβ and MYC and the suppression of Notch signaling pathway are likely the contributing factors to salinomycin’s cytotoxic effects. Taken together, this study suggests that salinomycin is a potential effective therapeutical agent for MB and warrants further investigation.
Acknowledgements
This work was supported by a project grant from the National Center for Research Resources (NCRR; P20 RR020151) and the National Institute of General Medical Sciences (NIGMS; P20 GM103505) from the National Institutes of Health (NIH). The contents of this report are solely the responsibility of the authors and do not necessarily reflect the official views of the NIH, NCRR, or NIGMS.
Conflicts of Interest
The author(s) confirm that this article content has no conflicts of interest.
References
- Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008; 3: 341-365.
- Hadjipanayis CG, Van Meir EG. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol Med. 2009; 15: 519-530.
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identiication of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009; 138: 645-659.
- Chapkin RS, Kim W, Lupton JR, McMurray DN. Dietary docosahexaenoic and eicosapentaenoic acid: emerging mediators of inlammation. Prostaglandins Leukot Essent Fatty Acids. 2009; 81: 187-191.
- Fuchs D, Daniel V, Sadeghi M, Opelz G, Naujokat C. Salinomycin overcomes ABC transporter-mediated multidrug and apoptosis resistance in human leukemia stem cell-like KG-1a cells. Biochem Biophys Res Commun. 2010; 394: 1098-1104.
- Fuchs D, Heinold A, Opelz G, Daniel V, Naujokat C. Salinomycin induces apoptosis and overcomes apoptosis resistance in human cancer cells. Biochem Biophys Res Commun. 2009; 390: 743-749.
- Kim JH, Chae M, Kim WK, Kim YJ, Kang HS, Kim HS, et al. Salinomycin sensitizes cancer cells to the effects of doxorubicin and etoposide treatment by increasing DNA damage and reducing p21 protein. Br J Pharmacol. 2011; 162: 773-784.
- Kim JH, Yoo HI, Kang HS, Ro J, Yoon S. Salinomycin sensitizes antimitotic drugs-treated cancer cells by increasing apoptosis via the prevention of G2 arrest. Biochem Biophys Res Commun. 2012; 418: 98-103.
- Kim KY, Yu SN, Lee SY, Chun SS, Choi YL, Park YM, et al. Salinomycin- induced apoptosis of human prostate cancer cells due to accumulated reactive oxygen species and mitochondrial membrane depolarization. Biochem Biophys Res Commun. 2011b; 413: 80-86.
- Kim WK, Kim JH, Yoon K, Kim S, Ro J, Kang HS, et al. Salinomycin, a p-glycoprotein inhibitor, sensitizes radiation-treated cancer cells by increasing DNA damage and inducing G2 arrest. Invest New Drugs. 2012; 30: 1311-1318.
- Zhou S, Wang F, Wong ET, Fonkem E, Hsieh TC, Wu JM, et al. Salinomycin: a novel anti-cancer agent with known anti-coccidial activities. Curr Med Chem. 2013; 20: 4095-4101.
- Ketola K, Hilvo M, Hyötyläinen T, Vuoristo A, Ruskeepää AL, Orešic M, et al. Salinomycin inhibits prostate cancer growth and migration via induction of oxidative stress. Br J Cancer. 2012; 106: 99-106.
- Lu D, Choi MY, Yu J, Castro JE, Kipps TJ, Carson DA. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci U S A. 2011; 108: 13253-13257.
- Zhang B, Wang X, Cai F, Chen W, Loesch U, Bitzer J, et al. Effects of salinomycin on human ovarian cancer cell line OV2008 are associated with modulating p38 MAPK. Tumour Biol. 2012; 33: 1855-1862.
- Keles GE, Berger MS, Lim R, Zaheer A, et al. Expression of glial ibrillary acidic protein in human medulloblastoma cells treated with recombinant glia maturation factor-beta. Oncol Res. 1992; 4: 431-437.
- Burger PC, Grahmann FC, Bliestle A, Kleihues P. Differentiation in the medulloblastoma. A histological and immunohistochemical study. Acta Neuropathol. 1987; 73: 115-123.
- Morrison LC, McClelland R, Aiken C, Bridges M, Liang L, Wang X, et al. Deconstruction of medulloblastoma cellular heterogeneity reveals differences between the most highly invasive and self-renewing phenotypes. Neoplasia. 2013; 15: 384-398.
- Tang X, Yao Y, Zhu J, Jin K, Wang Y, Mao Y, et al. Differential proliferative index of cancer stem-like cells in primary and recurrent medulloblastoma in human. Childs Nerv Syst. 2012; 28: 1869-1877.
- Zanini C, Ercole E, Mandili G, Salaroli R, Poli A, Renna C, et al. Medullospheres from DAOY, UW228 and ONS-76 cells: increased stem cell population and proteomic modiications. PLoS One. 2013; 8: e63748.
- Bernard F, Krejci A, Housden B, Adryan B, Bray SJ. Speciicity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development. 2010; 137: 2633-2642.
- Manoranjan B, Venugopal C, McFarlane N, Doble BW, Dunn SE, Scheinemann K, et al. Medulloblastoma stem cells: where development and cancer cross pathways. Pediatr Res. 2012; 71: 516-522.
- Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin Cancer Res. 2008; 14: 5314- 5317.
- Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006; 66: 7445-7452.
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004; 64: 7787-7793.
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004; 64: 7794-7800.
- He XM, Wikstrand CJ, Friedman HS, Bigner SH, Pleasure S, Trojanowski JQ, et al. Differentiation characteristics of newly established medulloblastoma cell lines (D384 Med, D425 Med, and D458 Med) and their transplantable xenografts. Lab Invest. 1991; 64: 833-843.
- Abouantoun TJ, Castellino RC, MacDonald TJ. Sunitinib induces PTEN expression and inhibits PDGFR signaling and migration of medulloblastoma cells. J Neurooncol. 2010; 101: 215-226.
- Abouantoun TJ, Macdonald TJ. Imatinib blocks migration and invasion of medulloblastoma cells by concurrently inhibiting activation of platelet- derived growth factor receptor and transactivation of epidermal growth factor receptor. Mol Cancer Ther. 2009; 8: 1137-1147.
- Park AK, Lee SJ, Phi JH, Wang KC, Kim DG, Cho BK, et al. Prognostic classiication of pediatric medulloblastoma based on chromosome 17p loss, expression of MYCC and MYCN, and Wnt pathway activation. Neuro Oncol. 2012; 14: 203-214.
- Stearns D, Chaudhry A, Abel TW, Burger PC, Dang CV, Eberhart CG. c-myc overexpression causes anaplasia in medulloblastoma. Cancer Res. 2006; 66: 673-681.
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009; 66: 1631-1646.
- Zweidler-McKay PA. Notch signaling in pediatric malignancies. Curr Oncol Rep. 2008; 10: 459-468.
- Huczynski A. Salinomycin: a new cancer drug candidate. Chem Biol Drug Des. 2012; 79: 235-238.
- Naujokat C, Fuchs D, Opelz G. Salinomycin in cancer: A new mission for an old agent. Mol Med Rep. 2010; 3: 555-559.
- Naujokat C, Steinhart R. Salinomycin as a drug for targeting human cancer stem cells. J Biomed Biotechnol. 2012; 2012: 950658.
- Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992; 11: 961-971.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Grifin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000; 26: 484-489.
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Grifin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000; 26: 484-489.
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001; 20: 3427-3436.Yochum GS, Cleland R, Goodman RH. A genome-wide screen for beta- catenin binding sites identiies a downstream enhancer element that controls c-Myc gene expression. Mol Cell Biol. 2008; 28: 7368-7379.
- Yochum GS, Cleland R, Goodman RH. A genome-wide screen for beta- catenin binding sites identiies a downstream enhancer element that controls c-Myc gene expression. Mol Cell Biol. 2008; 28: 7368-7379.
- Ntziachristos P, Lim JS, Sage J, Aifantis I. From Fly Wings to Targeted Cancer Therapies: A Centennial for Notch Signaling. Cancer Cell. 2014; 25: 318-334.
- Orian A, Delrow JJ, Rosales Nieves AE, Abed M, Metzger D, Paroush Z, et al. A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc Natl Acad Sci USA. 2007; 104: 15771-15776.
- Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999; 401: 86-90.
- Armelin HA, Armelin MC, Kelly K, Stewart T, Leder P, Cochran BH, et al. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984; 310: 655-660.
- Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD. Ampliication of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res. 1990; 50: 2347-2350.
- Herms J, Neidt I, Lüscher B, Sommer A, Schürmann P, Schröder T, et al. C-MYC expression in medulloblastoma and its prognostic value. Int J Cancer. 2000; 89: 395-402.
- Siu IM, Lal A, Blankenship JR, Aldosari N, Riggins GJ. c-Myc promoter activation in medulloblastoma. Cancer Res. 2003; 63: 4773-4776.
- Cappellen D, Schlange T, Bauer M, Maurer F, Hynes NE. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007; 8: 70-76.
- Eberhart CG. Three down and one to go: modeling medulloblastoma subgroups. Cancer Cell. 2012; 21: 137-138.
- von Bueren AO, Shalaby T, Oehler-Jänne C, Arnold L, Stearns D, Eberhart CG, et al. RNA interference-mediated c-MYC inhibition prevents cell growth and decreases sensitivity to radio- and chemotherapy in childhood medulloblastoma cells. BMC Cancer. 2009; 9: 10.
- Gilbertson RJ, Clifford SC. PDGFRB is over expressed in metastatic medulloblastoma. Nat Genet. 2003; 35: 197-198.
- Wang Y. Effects of salinomycin on cancer stem cell in human lung adenocarcinoma A549 cells. Med Chem. 2011; 7: 106-111.