1Department of Pediatrics, Hematology and Oncology, University Medical Center of the Georg- August- University Goettingen, Germany
2Schaller Research Group at the University of Heidelberg and the German Cancer Research Center, Division of Molecular Mechanisms of Tumor Invasion, German Cancer Research Center (DKFZ), Germany
3Department of Molecular Pharmacology, Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Israel
4FluoronGmbH, Magirus-Deutz-Straße, Germany
5Musculoskeletal Research Laboratopry, Rambam – Health Care Campus, Orthopaedics Department, Israel
#Equal Contribution of Leo Veenman and Julia Bode
*Corresponding author: Moshe Gavish, Department of Molecular Pharmacology, Ruth and Bruce Rappaport Faculty of Medicine, Technion-Israel Institute of Technology, Ephron Street, P.O.B. 9649, Bat-Galim, Haifa 31096, Israel
Received: August 04, 2014; Accepted: October 14, 2014; Published: October 15, 2014
Citation: Bode J, Veenman L, Vainshtein A, Kugler W, Rosenberg N and Gavish M. Modulation of Gene Expression Associated with the Cell Cycle and Tumorigenicity of Glioblastoma Cells by the 18 kDa Translocator Protein (TSPO). Austin J Pharmacol Ther. 2014; 2 (10).1053. ISSN: 2373-6208.
After PK 11195 (25 μM) exposure (24 and 48 hours) and permanent TSPO silencing by siRNA to U118MG glioblastoma cells, microarray analysis of gene expression, followed up by validation with real time polymerase chain reaction (RT-PCR), showed effects on gene expression related to the cell cycle. Other affected genes are related to apoptosis, oxidative stress, immune responses, DNA repair, and membrane functions, including adhesion and transport. In relation to post transcriptional and post translational effects, TSPO ligand and TSPO knockdown affected gene expression for many short nucleolar RNAs. Applying a 2-fold, cut off to micro array analysis revealed that 24 and 48 hours of PK 11195 exposure affected respectively 128 and 85 genes that were also affected by TSPO silencing. Taking a 2.5 fold, cut off, only gene expression of v-FOS was found to be modulated by each of these TSPO manipulations. Analyzing the effects of TSPO knockdown and PK 11195 exposure, RT-PCR indicated that 5 genes related to the cell cycle are under TSPO control. Focusing further on the cell cycle: 1) TSPO silencing increased numbers of cells in the S (to 321%) and G2/M (to 131%) phases, and caused a decrease in the G0/G1 phase (to 69%); 2) 24 hours of PK 11195 exposure caused decreases in the S (to 81%) and G2/M (to 94%) phases, and an increase in the G0/G1 (to 112%) phase; 3) 48 hours of PK 11195 exposure caused decreases in the S (to 70%) and G0/G1 (to 96%) phases, and an increase in the G2/M (to 124%) phase. Thus, TSPO appears to provide for regulation of the cell cycle via specific modulation of gene expression for key gene products in these glioblastoma cells.
Keywords: Glioblastoma; 18 kDa Translocator protein (TSPO); Gene expression; FOS; Cell cycle; siRNA knockdown; PK 11195; Brain tumor
BRCA1: Breast Cancer 1, Early Onset; CDC2: Cell Division Cycle 2; CDC6: Cell Division Cycle 6; EGR1: Early Growth Response 1; FGIN-1-27: N, N-Dihexyl-2-(4-fluorophenyl)Indole-3-Acetamide; GBM: Glioblastoma Multiforme; MCM2: Minichromosome Maintenance Complex Component 2; MCM4: Minichromosome Maintenance Complex Component 4; PK 11195: 1-(2-Chlorophenyl)- N-methyl-N-(1-methylpropyl)-3-Isoquinolinecarboxamide; PPIX: Protoporphyrin IX; RANBP3: Ran-Binding Rotein 3; Ro5-4864: Benzodiazepine Ro5 4864 (4´- chlorodiazepam); ROS: Reactive Oxygen Species; RT-PCR: Realtime Polymerase Chain Reaction; TSPO: 18 kDa Translocator Protein; v-FOS: v- fos FBJ murine osteosarcoma viral oncogene homolog
Glioblastoma (GBM) presents the most aggressive form of malignant glioma, including spread and destruction of tissue throughout the brain [1,2]. Moreover, current treatment regimens only providemarginalimpact on patient survival [3]. Survival rate after diagnosis of GBM typically is less than 6 months. GBM presents 80% of the malignant gliomas [4-6]. The challenge is to understand and arrest the cellular processes contributing to the malignancy of glioblastoma. [7]. Previous studies by us have shown that glioblastoma cells could be transformed to even more malignant strains due to silencing of the 18 kDa translocator protein (TSPO)by genetic manipulation [8-12]. Thus, it is important to understand the role ofthe TSPO in malignancy [13,14]..
TSPO can be found in normal glial cells as well as healthy peripheral tissues such as the heart, lung, kidney, adrenal, and reproductive organs [14,15].. In addition, TSPO expression typically is enhanced in tumorigenic tissues and cells.In the mitochondria, TSPO can be found in association with several proteins that are related to various functions such as cell metabolism, programmed cell death, heme metabolism, ROS generation, ATP production, and steroidogenesis [11,16-20]. The association of the TSPO with the cell nucleus is thought to take part in regulation of gene expression by the TSPO [21,22]. Synthetic TSPO ligands include the benzodiazepine Ro5 4864 (4´- chlorodiazepam), as well as the isoquinoline carboxamide derivate PK 11195,and have been used to study the functions of the TSPO [23,26]. The TSPO has been implicated in various functions, for example,programmed cell death, steroidogenesis, mitochondrial respiration, reactive oxygen species (ROS) generation, regulation of the mitochondrial membrane potential (Δψm), inflammation, effects on the immune and phagocytic host-defense response, microglial activation, ischemia, cell growth and differentiation, cancer cell proliferation, regulation of the cell cycle, and mental and neuropathological disorders [14,17,20,25-29]. Several of these functions have implications for TSPOinvolvement in tumorigenicity. We have previously described the effect of TSPO knockdown by siRNA as well as treatments with the specific TSPO ligands PK 11195 and Ro5 4864 on programmed cell death of U118MG glioblastoma cells [11,12,18,30,31]. In addition, enhanced TSPO expression in cancer cells correlates with their proliferation [9,32]. Furthermore, TSPO knockdown as well as application of the TSPO ligand PK11195reduced adhesion and enhanced migratory characteristics of glioma cells in cell culture in vitro and increased invasiveness in vivo in animal models [2]. Microarray analysis of gene expression suggested that the TSPO may regulate gene expression related to tumorigenicity of U118MG cells [22]. The influence of TSPO knockdown on the cell cycle of glioblastoma cells was for example studied on the rat C6 cell line [9]. In that study the TSPO knockdown resulted in a lower percentage of knockdown clones in the G0/G1 phase and a larger percentage of the same clones in the G2/M phases in comparison to the control. It also was shown that synthetic TSPO ligands, such as FGIN-1-27, PK 11195,and Ro5 4864,dose-dependently inhibited cell proliferation of via an arrest of the G0/G1 phase in various human cancer cells [33-36]. Thusfar, none of these studies proposed a mechanism whereby TSPO may be able to affect the cell cycle, nor do they present an explanation why opposing, apparently contradictory results can be found.
To better understand the involvement of the TSPO in regulation of the cell cycle due to TSPO modulations, we performed microarray analysis of gene expression.Our methodology includes study of the effects of the synthetic TSPO ligand PK 11195 applied for 24 and 48 hours in comparisons with the effects of permanent TSPO knockdown by siRNA introduced by stable transfection. By this approach we consider the likelihood that effects that are common to these different methods indeed are due to effects on the TSPO, while effects unique to either one of these methods do not necessarily relate directly to the TSPO. After screening the general effects on gene expression in our paradigm, we focussed ongene expression associated with the cell cycle, and also on actual changes in the cell cycle phasesin these complementary experimental models.
Materials required for our cell cultures were obtained from Biological Industries (Beit Ha’emek, Israel), Biochrom (Berlin, Germany), and Lonza (Basel, Switzerland). Oligonucleotides for real time polymerase chain reaction (RT–PCR) were from IBA (BioTAGnology, Göttingen, Germany) and SYBR Green Mastermix from Applied Biosystems (Foster City, CA). Materials for microarray analysis were from Agilent (Life science group, Penzberg, Germany) and Affymetrix (Santa Clara, CA). TRIzol reagent used for RNA extraction of microarray samples was purchased Invitrogen (Darmstadt, Germany). 1-(2-Chlorophenyl)-N-methyl-N-(1- methylpropyl)-3-isoquinolinecarboxamide (PK 11195) was obtained from Alexis (Switzerland).
Cells of the human glioblastoma cell line U118MG were cultured at 37oC in an atmosphere of 5% CO2 and 90% relative humidity. U118MG wild type (WT) cells were cultured in MEM-EAGLE cell culture mediumsupplemented with 10% FBS, 2% glutamine, and 0.05% gentamyc in (50 mg/ml). We applied stable transfection with siRNA to silence TSPO expression in U118MG cells (TSPO knockdown) and also prepared sham control scrambled siRNA cells (Scr control) as described before [12]. Expression of TSPO was routinely assessed using RT-PCR and Western blot, as described by us in detail previously [11,12]. In the present study, RT-PCR assays showed that gene expression of the TSPO protein was reduced to 20% of the levels detected in Scr control cells (n = 6, p < 0.01, for each assay), similar to what was described in previous studies [11,12,31]. Forthe assays of the present study (flow cytometry for cell cycle analysis; and RNA extraction for microarray and RT-PCR), TSPO knockdown cells, the Scr control cells, and the WT cells were grown until they reached 90% confluence. Cells were collected by trypsinization and centrifugation, as described previously [9,11,12,22], to study the effects of our treatments on gene expression and the cell cycle.
WT U118MG cells (1 × 106) were seeded in 75 cm2 cell culture flasks and allowed to acclimatize overnight. Then the cells were exposed to 25 μM of PK 11195 (Alexis, Switzerland). The exposure times to PK 11195 were 24 and 48 hrs. Flow cytometry and RNA extraction were executed immediately after the PK 11195 exposures. Stock solution of PK 11195 with a concentration of 20 mM was prepared in 100% ethanol. For exposures, final concentration of PK 11195 was 25 μM in the culture medium and final concentration of ethanol was 1%. Control cells were exposed to vehicle i.e. 1% ethanol final concentration [30]. The concentration of 25 μM of PK 11195 was previously found in other studies to be very effective and consistent, providing effects that were similar to those of TSPO knockdown by genetic manipulation in those studies [2,11,12,18,22,30,31].
For microarray analysis of gene expression, 3 × 104 U118MG cells (Scr control and siRNA TSPO knockdown) were seeded and grown in cell culture flasks (75 cm2), until they reached 90% confluency as described above. For these assays, also WT U118MG cells were seeded, cultured, and exposed to PK 11195 for 24 and 48 hrs as described above. Experiments were repeated at least three times, and each experiment was done in triplicates. Performing of RT-PCR was performed as described before [2]. Cells were collected by trypsinization and in this way 3 samples of 3 × 105 cells were obtained per experimental group, each sample coming from a separate flask. Cell pellets were prepared by centrifugation (1000 × g, 7 min, 20°C). The RNA isolation was done according to the “TRIzol RNA extraction protocol” (Invitrogen, Darmstadt, Germany). The RNA pellet was dissolved in 100 μl RNase free water and mixed by repetitive pipetting. For checking RNA quality and quantity, 2 μl of each sample was used. Measurements of the 28S:18S ratios to determine the integrity of RNA samples were done using Agilent 2100 bioanalyzer (Agilent Technologies, Genomics, Santa Clara, CA), as described previously [11,22,37,38]. The ‘‘Low RNA Input linear Amplification Kit Plus, two color’’ (Agilent, 5188-5340) and the ‘‘RNA Spike-In Kit’’ (Agilent, 5188-5279) were used for cDNA synthesis and in vitro transcription according to the manufacturer’s recommendations. Quantity and dye incorporation rates of the amplified cRNAs were determined using the NanoDrop ND-1000 UV-VIS Spectrophotometer version 3.2.1 (NanoDrop Technologies, Wilmington, DE). cDNA was labeled for microarray hybridization with the help of an in vitro transcription (IVT) labeling reaction. The IVT labeling was performed with the help of Affymetrix GeneChip® IVT Labelling Kit (Affymetrix, High Wycombe, UK). Fragmentation and hybridization of labeledamino-allyl-RNA (aRNA) GeneChip® Human Exon 1.0 ST Array (Affymetrix) was performed in the DNA Microarray Facility at the Medical Faculty of the Georg-August- University of Göttingen [37,39-42]. Briefly, Affymetrix Whole Transcript chips were used and hierarchical gene cluster analysis was applied. The raw intensity data were normalized with quantile normalization. Differentially expressed genes were identified by an ANOVA fixed effects model. Adjusted p-values were obtained by the Benjamini-Hochberg method to control the false discovery-rate [43,44]. To further avoid false positives in our interpretations, we considered only 2- fold differences and higher between gene expression of detected genes in Scr and TSPO knockdown cells, and between vehicle control and PK 11195 exposure detected with our microarray analysis. For the present study, microarray assays were applied to U118MG TSPO knockdown cells and their Scr control cells, and to WT U118MG cells exposed for 24 hrs and 48 hrs to PK 11195 and their vehicle controls.
To verify the results of the screening by microarray assay, as another measure to avoid false positives, RT-PCR was applied as described [2,22]. For the present study we applied RT-PCR to genes coding for proteins with relatively well defined functions in the cell cycle that were firstly detected by microarray applied to U118MG TSPO knockdown cells in comparison to Scr control cells (Table 1), applying the criterion of a 2-fold cut off. For these genes we also applied RT-PCR to U118MG cells exposed to PK 11195 for 24 hrs and 48 hrs, if they showed significant differences with the microarray analysis (Table 1). RNA extraction was applied according to the instructions provided with the kit (RNeasy® Mini Kit, Qiagen, Germany). The concentrations of RNAs were determined by NanoDrop™ measurement. Reverse transcription of RNA was performed with iScript cDNA Synthesis Kit (BioRad, Hercules, CA), according to the instructions of the manufacturer. RT-PCR was performed using 10 ng target cDNA and 3 pmol of each primer in question (see list below) in SYBR-GreenMix (SYBR Green, Applied Biosystems, Foster City, CA). The reaction was carried out with a final volume of 10 μl in 384 well optical plates. For each sample, triplicates were performed and water was taken as control. 18S was chosen as an internal standard because its expression is stable irrespective of treatment of cells [45]. The constancy of the cycle threshold (ct) of 18S can be measured and analyzed after 12 cycles for all experiments. The reaction was carried out at the ABI PRISM 7900HT Fast Real Time PCR System (Applied Biosystems, Foster City, CA) with 40 cycles of 15 seconds at 95°C, and one cycle of one minute at 60°C followed by the measurement of the dissociation curve. Data analysis of RT-PCR was performed using SDS 2.4 software (Applied Biosystems, Foster City, CA). For this purpose, the threshold was adjusted to the beginning of exponential fluorescence increase in a logarithmic plot of fluorescence over the amount of cycles. This was done to eliminate the background noise of fluorescence, as described previously [45]. The point of intersection between fluorescence curve and threshold defines the ct-value (cycle threshold) of the respective gene in the respective sample i.e. the time point at which the chosen threshold was attained defined the level of change in gene expression. The mean of the triple ct-values was normalized to the internal standard creating the Δct-value. The ct-value of the internal standard 18S did not differ between all analyses of gene expressions at 12 cycles, indicative of the consistency of the RT-PCR assays for our analyses. The difference between Δct-values of TSPO knockdown or PK 11195 treated cells and their respective control cells defines the ΔΔct-value. Due to the exponential relation of the DNA amount to the respective cycle, the normalized difference of mRNA that was present in the original samples is described by 2-ΔΔt. This value represents the fold change of expression induced by the treatment [45].
Primer sequences used for realtime-PCR analysis:
BRCA1 Forw 5´-TTCCAGATCCACAAGCCCAAC-3´
Rev 5´-GGCACTATTCTCTGATGACCCG-3´
CDC2 Forw 5´-TGATCCAGCCAAACGAATTTC-3´
Rev 5´-GCTACATCTTCTTAATCTGATTGTCCAA-3´
CDC6 Forw 5´-CCGTAACCTGTTCTCCTCGTGT-3´
Rev 5´-TCTTGCCTTGCTTTGGTGGA-3´
Claspin Forw 5´-GGATCCGCCGCCACCATGACAGGCGAGGTGGGT-3´
Rev 5´-TCTAGACTCGAGGCTCTCCAAATATTTG-3´
CyclinE2 Forw 5´-TGTTGGCCACCTGTATTATCTGG-3´
Rev 5´-ATCTGGAGAAATCACTTGTTCCTATTTC-3´
CyclinG2 Forw 5´-GCTGAAAGCTTGCAACTGCCGACTC-3´
Rev 5´-GGTATCGTTGGCAGCTCAGGAAC-3´
EGR1 Forw 5´-ACCGCAGAGTCTTTTCCTGACA-3´
Rev 5´- TTGGTCATGCTCACTAGGCCAC-3´
KIAA Forw 5´-AACATGGTGCGGACTAAAGCA-3´
Rev 5´-CCTCGATGAAACTGATGTCGAA3´
MCM2 Forw 5´-AGACGAGATAGAGCTGACTG-3´
Rev 5´-CACCACGTACCTTGTGCTTG-3´
MCM4 Forw 5´-CCTTCAGAGATTTCTTCAGCG-3´
Rev 5´-TCACCAATAACATTAATCTCCC-3´
v-FOS Forw 5´-TGGCGTTGTGAAGACCATGA-3´
Rev 5´-CCCTTCGGATTCTCCTTTTCTC-3´
RANBP3 Forw 5´-ACAGCAAGCACTGACTGTGG-3´
Rev 5´-GGCACTGGCCTGAATTAAAA-3´
18S Forw 5´-AACTTTCGATGGTAGTCGCCG-3´
Rev 5´-GGATGTGGTAGCCGTTTCTCAG-3´
For cell cycle analysis, Fluorescence Assisted Cell Sorting (FACS) analysis of the cell cultures of our study was performed each time by 3 independent experiments each done in triplicate. In brief,cells were harvested by trypsinization.Cells were pelleted by centrifugation, resuspended, and fixed in 500 μl of Nicoletti buffer [0.1% (w/v) Sodium Citrate, 0.1% (v/v) of TritonX-100 and 50μg/ml of propidium iodide (PI)]. The cell suspension was vortexed and incubated for 60 min at 4°C in the dark. Cells were analyzed immediately for PI labeling using the FACScan flow cytometer (Becton Dickinson, Heidelberg, Germany). For each measurement, 10,000 cells were counted. Dot plots and histograms were analyzed by Cell-Quest Pro software (BD Biosciences, Heidelberg, Germany).
All assays were at least 3 independent experiments, each done in triplicate (total n ≥ 9). Error bars represent the standard deviation (SD). For statistical analysis paired Student´s t-Test was applied as appropriate. Statistical significance is presented in the following manner: * p < 0.05, ** p < 0.01, and *** p < 0.001.
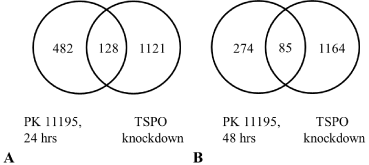
Applying a 2.0 fold change cut off for our micro array screening, Venn diagrams present the number of genes that underwent significant changes in expression common for TSPO knockdown cells and WT cells exposed to PK 11195 for 24 hrs and 48 hrs, respectively (Figure 1) (see also the appendices 1, 2, 3). When WT U118MG cells were exposed for 24 and 48 hours to 25 μM of PK 11195, significant changes in expression detected in this way were respectively 610 and 359 genes (Figure 1A and B). Compared to U118MG Scr control cells, knockdown of TSPO with siRNA resulted in significant changes in gene expression levels for 1249 genes (Figure 1) [22]. Furthermore, the Venn diagrams (Figure 1) demonstrated an overlap of 128 genes having their expression changed by TSPO knockdown as well as 24 hrs of PK 11195 exposure (Figure 1A). With the same approach, WT cells exposed to PK 11195 for 48 hrs and TSPO knockdown cells presented an overlap of 85 genes (Figure1B).
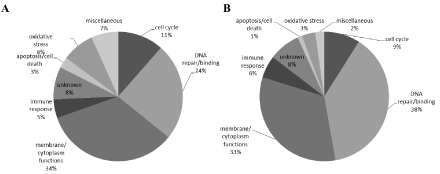
We also considered the functional nature of the genes that showed significant 2 fold changes effected by TSPO knockdown as well as by PK 11195 exposure for 24 hours and 48 hours, as compared to their respective controls. These genes were categorized into functional entities that included: membrane and cytoplasm functions (transport, cytoskeleton, and adhesion), effects on DNA, the cell cycle, oxidative stress, immune response, and cell death (Figure 2). The proportions of the overlaps between the groups of genes found with these comparisons were about the same for most of the functional categories (Figure 2). One exception was the effect on expression of genes related to oxidative stress, which constituted 8% of the total population of affected genes found to be affected both in TSPO knockdown cells and cells exposed for 24 hours to 25 μM of PK 11195. However, such genes formed only 3% of the total overlap of changes in gene expression between TSPO knockdown cells and cells exposed for 48 hours to 25 μM of PK 11195. Another functional entity showing considerable changes in gene expression both in TSPO knockdown cells and cells exposed for 24 hours to 25 μM of PK 11195 was DNA repair and binding to DNA, presenting an overlap which constituted 24% of the total population of affected genes in this comparison. This increased to 38% of the total overlap of changes in gene expression between TSPO knockdown cells and cells exposed for 48 hours to 25 μM of PK 11195. Furthermore, a general observation, our micro array analysis showed that gene expression affected by TSPO knockdown and TSPO ligand exposure corresponded to functions typically attributed to the TSPO [14,22,26].
Subsequently, we focused on the effects of TSPO knockdown and PK 11195 exposure (24 hrs and 48 hrs) on gene expression related to cell cycle, presented by microarray screening (Table 1). Table 1 shows this screened gene expression related to the cell cycle, including the direction of the changes (+ or - ) and the magnitudes of the changes. Table 1 also presents the specific functions in the cell cycle related to the genes in question.
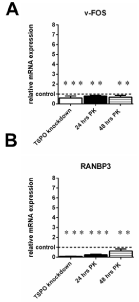
Table 1 also presents v-FOS and RANBP3 gene expression, which showed the biggest changes in expression in the present study.With a cut off of a 2.5 change, only v-FOS FBJ murine osteosarcoma viral oncogene homolog (v-FOS) showed a change in gene expression due to TSPO knockdown, as well as both the 24 hrs and 48 hrs exposures to PK 11195 (Table 1). In more detail, v-FOS expression levels were decreased to 0.2 fold in U118MG TSPO knockdown cells, to 0.3 fold after 24 hrs as well as 48 hrs of PK 11195 exposure of WT cells (Appendices 1, 2, 3, and Table 1). With this 2.5 cut off applied, Ran-binding protein 3 (RANBP3) also showed a change in gene expression with TSPO knockdown and with 24 hrs exposure to PK 11195 (Table 1), but not with 48 hrs exposure to PK 11195.In more detail, changes in expression levels of RANBP3 were 0.1 fold for TSPO knockdown, 0.4 fold for 24 hrs of PK 11195 exposure, and 0.5 fold for 48 hrs of PK 11195 exposure (Appendices 1, 2, 3, and Table 1). It has been reported that the v-FOS gene either encodes or induces an activator of transcription that recognizes specific sequences in promoters [46]. RANBP3 is a cofactor for Crm1-mediated nuclear protein export [47].
To verify the effects of TSPO knockdown and PK 11195 exposure (24 hrs and 48 hrs) on gene expression related to cell cycle (Table 1, appendices 1,2,3), RT-PCR was subsequently applied (Figures. 3-5). In addition, RT-PCR was subsequently applied to the v-FOS and RANBP3genes (Figure 6), which had displayed more than 2.5-fold changes in expression as observed with microarray analysis of TSPO knockdown cells, as presented above. Figures 3, 4, and 5 show these RT-PCR results:
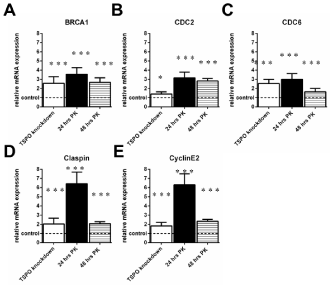
Figure 3 presents genes that show changes due to TSPO knockdown as well as both 24 hours and48 hours of PK 11195 exposure,as obtained with RT PCR: Breast cancer 1, early onset (BRCA1), Cell division cycle 2 (CDC 2), Cell division cycle 6 homolog (CDC6), Claspin homolog (Claspin), and CyclinE2 (CyclinE2). In more detail:
BRCA1 was upregulated to 257% ± 73 after TSPO knockdown (Control 100% ± 15), to 355% ±73 after 24 hrs of PK 11195 exposure (Control 100% ± 22), and to 267% ±50 after 48 hrs of PK 11195 exposure (Control 100% ± 13) (Figure 3A).
CDC 2 was upregulated to 140% ± 24 after TSPO knockdown (Control 100% ± 28), to 316% ± 62 after 24 hrs of PK 11195 exposure (Control 100% ± 20), and to 283% ± 27 after 48 hrs of PK 11195 exposure (Control 100% ± 5) (Figure 3B). CDC6 was upregulated to 255% ± 45 after TSPO knockdown (Control 100% ± 14), to 300% ± 63 after 24 hrs of PK 11195 exposure(Control 100% ± 21), and to 164% ± 37 after 48 hrs of PK 11195 exposure (Control 100% ± 28) (Figure 3C).
Claspinwas upregulated to 204% ±63 after TSPO knockdown (Control 100% ± 24), to 643% ±126 after 24 hrs of PK 11195 exposure (Control 100% ± 19), and to 207% ±23 after 48 hrs of PK 11195 exposure (Control 100% ± 18) (Figure 3D). CyclinE2 was upregulated to 183% ± 38 after TSPO knockdown (Control 100% ± 28), to 630% ± 120 after 24 hrs of PK 11195 exposure (Control 100% ± 18), and to 244% ± 20 after exposure for 48 hrs after 48 hrs of PK 11195 exposure(Control 100% ± 16) (Figure 3E).
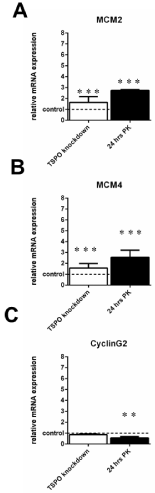
Figure 4 presents genes that show changes due to TSPO knockdown as well as 24 hours of PK 11195 exposure, but have nosignificant changes after 48 hours of PK 11195 exposure.After TSPO knockdown and after exposure for 24 hrs of PK 11195, the gene expression levels for minichromosome maintenance complex component 2 (MCM2), minichromosome maintenance complex component 4 (MCM4), and CyclinG2 were changed more than 2-fold by both these two treatments of U118MG cells, as firstly seen with microarray (Table 1). Applying RT-PCR, Figure 4A shows that gene expression of MCM2 in TSPO knockdown cells was upregulated to 164% ± 53 (Control 100% ± 36) and after 24 hrs of PK 11195 exposure to 274% ±8 (Control 100% ±13).The gene expression for MCM4 was upregulated to 157% ± 41 in TSPO knockdown cellsand after 24 hrs of PK 11195 exposure to 256% ± 65 (Control 100% ± 2) (Figure 4B). Gene expression of CyclinG2 (CyclinG2) was downregulated to 86% ± 7 after TSPO knockdown (Control 100% ± 31), and to 57% ± 13 after 24 hrs of PK 11195 exposure (Control 100% ± 6) (Figure 4C).
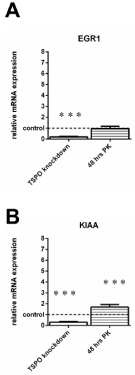
Figure 5 presents genes that show changes due to TSPO knockdown as well as 48 hours of PK 11195 exposure, but have nosignificant changes after 24 hours of PK 11195 exposure. Figure 5 shows the RT-PCR results for early growth response 1 (EGR1) and KIAA (KIAA 1524)in TSPO knockdown cells and after exposure of PK 11195 for 48 hrs, which a priori showed more than two fold changes in gene expression as measured with microarray (Table 1). The gene expression of EGR1 was significantly downregulated in TSPO knockdown cells to 20% ± 5 (Control 100% ± 18), but not so after 48 hrs of PK 11195 exposure(to 96%± 22) (Control 100% ± 33) (Figure5A). Furthermore, KIAA was downregulated to 30% ± 7 after TSPO knockdown (Control 100% ± 21)and upregulated to 169% after PK 11195 exposure for 48 hrs (Control 100% ± 25) (Figure 5B).
Interestingly, as mentioned above, v-FOS and RANBP3showed the biggest changes in gene expression with TSPO knockdown in the microarray analysis (Table 1). RT-PCR showed that TSPO knockdown led to a decrease in v-FOS expression to 59% ± 21 (Control 100% ± 15)and after 24 hrs and 48 hrs of exposure to PK 11195 to downregulations to 80%± 7and 66%± 21, respectively (Control 100% ± 27) (Figure 6A). Furthermore, TSPO knockdown led also to a decrease in RANBP3expression to 10% ± 2 (Control 100% ± 3) and decreases in expression to 24% ± 24and 62% ± 22(Control 100% ± 21) after PK 11195 exposure for 24 hrs and 48 hrs, respectively, as seen with RT-PCR (Figure 6B).
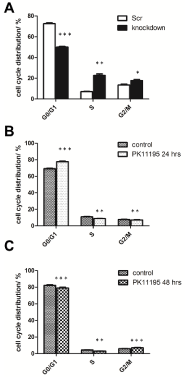
Cell cycle analysis / distribution of U118MG TSPO knockdown cells and PK 11195 (24 and 48 hrs) exposed U118MG cells compared to control cells was performed by FACS analysis. TSPO knockdown led to a significant reduction in the proportion of the number of cells in the G0/G1 phase [from 72.66% ± 0.80to 49.99%± 0.64,***p< 0.001], an increase in the proportion of the number of cells in the S phase [from 7.08% ± 0.33to 22.75%± 1.44,**p< 0.01], and an increase in the proportion of the number of cells in the G2/M phase [from 13.53% ± 0.92to 17.72%±1.05, *p < 0.05] (Figure 7A).
PK 11195 exposure for 24 hrs led to an increase in the proportion of thenumber of cells in the G0/G1 phase [from 69.14% ± 0.57to 77.60%± 0.68 (***p < 0.001)]and decreases in the proportions of the number of cells in theS phase [from 11.02% ± 0.53to 8.91%±0.21, **p< 0.01]and the G2/ M phase [from 7.62% ± 0.47to 7.16%± 0.33, * p< 0.05] (Figure 7B).
U118MG exposure to PK 11195 for 48 hrs led to smalldecreases in the proportions of the number of cells in the G0/G1 phase [from 82.19% ± 0.86to 79.00%± 1.08, ***p< 0.001] and in the S phase [from 4.28% ± 0.2to 3.0% ± 0.24, **p< 0.01], and an increase in the proportion of thenumber of cells in the G2/M phase [from 5.86% ± 0.24to 7.00%± 0.50, ***p<0.001] (Figure 7C).
Severalprevious studies have suggested that the TSPO is involved in glioblastoma tumorigenicity [26,32,48]. For example knockdown studies of TSPO, as well as application of TSPO ligands, have indicated pro cell death functions of the TSPO that include promotion of collapse of the mitochondrial membrane potential (Δψm), Reactive Oxygen Species (ROS) generation at mitochondrial levels, cardiolipin oxidation, leading to programmed cell death [9,12,16-20,30,32,49]. Furthermore, in cell culture, TSPO knockdown enhances anchorage independent cell proliferation (i.e. suspended in agar), reduces cell adhesion, and enhances cell migration of glioma cells [2,8,9]. Finally, in vivo studies in whole animal models have shown that TSPO knockdown in cancer cells causes enhanced glioblastoma tumor proliferation in the brain [2] and reduces survival rate of the host animals [2,8,9]. Nonetheless, a complete understanding of TSPO involvement in tumorigenicity is not simple. Part of this enigma is that the TSPO is involved in various and numerous functions [14,22,26]. We consider the possibility that TSPO’s involvement in regulation of gene expression may at least partly explain TSPO’s association with these functions.
We are driven by the realization that the TSPO presents a central component for processes related to the host defense response [17,26]. Thus, to better understand how TSPO can be involved in various and numerous functions, in particular related to tumorigenesis of glioblastoma, we studied TSPO involvement in gene expression. In particular, we selected 24 hours and 48 hours of exposure to the specific TSPO ligand PK 11195, at a concentration of 25 μM. This concentration of PK 11195 is known to inhibit TSPO functions,including programmed cell death in U118MG cells induced by various agents [2,12,30,31]. In addition, we applied stable knockdown of TSPO, which presents a permanent inhibition of TSPO function, including enhanced tumorigenicity and attenuation of programmed cell death in U118MG and other cell lines, an effect that can also be induced by TSPO ligands [2,8-12,18,19,22,30,31]. We consider that differences in the outcomes regarding gene expression following 24 and 48 hours of exposure to 25 μM of PK 11195, and stable TSPO knockdown may be due to the duration of changes in TSPO function (i.e 24 hrs., 48 hrs, and permanent).Indeed, we found that 24 hrs of PK 11195 (25 μM) exposure affected more genes than48 hours of exposure to PK 11195 (25 μM) (610 genes versus 359 genes). TSPO knockdown by stable siRNA transfection affected 1249 genes (when applying a minimum of a 2.0 fold, cut off), which was much more than found with 24 and 48 hours of PK 11195 exposure, applying the same cut off. When considering the overlapping populations of genes affected by the differing TSPO manipulations, also the number of genes affected both by 48 hours of PK 11195 (25 μM) and TSPO knockdown was considerably smaller than the number of genes affected both by 24 hrs of PK 11195 (25 μM) and TSPO knockdown (85 genes vs. 128 genes).
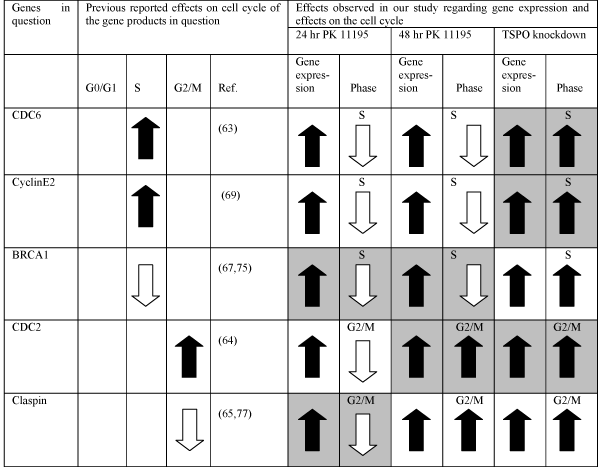
As regulation of the cell cycle is a major component for tumorigenesis we paid extra attention to the group of genes related to this cell function. Previous studies applying TSPO knockdown and various TSPO ligands already suggested TSPO involvement in the cell cycle [9,34-36,50,51]. The present study applied the approach of determining by microarray and RT-PCR which genes for proteins related to the cell cycle are under the control of the TSPO. With the present study we found that TSPO knockdown by siRNA, as well as exposure to 25 μM of the TSPO ligand PK 11195 for 24 hours and 48 hours, significantly modulates gene expression for several proteins associated with the cell cycle (Table 1). These individual proteins each regulate one or more of the several phases of the cell cycle. Details how these proteins may regulate various aspects of the cell cycle have been studied intensively by several groups [52-57]. As these genes were affected by TSPO knockdown as well as by exposure to PK 11195 we assume that such genes actually may be under the control of the TSPO. Genes that were affected only by TSPO knockdown or only by exposure to PK 11195 were not further scrutinized in the present study. They are however in the appendices 1, 2, and 3 added to this work and a previous work [22].
In table 2 we summarize gene expression related to the cell cycle that is affected by 24 and 48 hours of exposure to PK 11195, and by TSPO knockdown, as indicated by microarray as well as by RT-PCR. The following 5 genes, CDC6, CyclinE2, BRCA1, CDC2, and Claspin indicated in Table 2, fulfill these criteria i.e. they displayed changes after all three TSPO manipulations.We were also interested whether these changes in gene expression actually match changes in the cell cycle. In Table 2 we presentcorrelations between changes in gene expression and effects on the cell cycle following application of PK 11195 and of TSPO knockdown by siRNA. It is known that the proteins coded by CDC6 and CyclinE2 affect the S phase positively [53,58- 61,65] The protein coded by BRCA1 affects the S phase negatively [56,64]. CDC2 and Claspin code for proteins that affect the G2/M phase positively and negatively, respectively [54,55,66,67]. In this way, in TSPO knockdown cells, the upregulation of CDC6, CyclinE2, and CDC2 match well the upregulation of the S and G2/M phase (and consequently the downregulation of the G0/G1 phase in these cells) (Table 2). In cells exposed to PK 11195 for 48 hours, upregulation of the genes BRCA1 and CDC2 match well the downregulation of the S phase and the upregulation of the G2/M phase, respectively (Table 2). In cells exposed to PK 11195 for 24 hours, the upregulation of BRCA1 and Claspin match well the downregulation seen for both the S phase and the G2/M phase, respectively (and consequently the upregulation of the G0/G1 phase in these cells) (Table 2). Thus, while 24 and 48 hours of exposure to PK 11195 as well as TSPO knockdown all upregulate the expression of these 5 genes of CDC6, CyclinE2, BRCA1, CDC2, and Claspin (Figs. 3-5, and Table 1,2) the net functional consequences under these three conditions are not similar (Figure 7, and Table 2). In particular the functional effects of 24 hours of PK 11195 exposure and TSPO knockdown appear to be the exact opposite, while 48 hours of PK 11195 shows partial similarities with both these treatments. We considered the possibility that quantitative differences in changes in gene expression might explain the different functional outcomes, however, no clear cut association in magnitude of gene expression incorrelation with different changes in the cell cycle could be discerned (Figures 3-5, an d Table 2). As a consequence, we considered the possibility that pretranscriptional and posttranslational effects may play a role.
With this approach, we found that TSPO ligands and TSPO knockdown also affect gene expression of numerous non-coding genes, including small nucleolar RNAs (snRNAs) (Table 3) (see also appendices 1,2, and 3). Small nucleolar RNA (snoRNAs), function as a ribonucleoprotein particle (RNP) in the nucleolus. Certain snoRNAs have been reported to act as either oncogenes or tumor suppressors in vitro [68]. The central functions of snoRNAs have long been believed to modify, mature, and stabilize rRNAs. These post-transcriptional modifications are important for the production of efficient and accurate ribosomes [69]. However, snoRNAs can also partake in modifications of snRNAs that mediate mRNA splicing [70]. Furthermore, snoRNA transcripts serve as the precursors of miRNA-like small RNAs and the regulators of alternative splicing [71,72]. Interestingly, progressively larger numbers of such noncoding genes are affected by 24 hours of PK 11195 exposure, 48 hours of PK 11195 exposure, and TSPO knockdown by siRNA, respectively (Table 3) (see also appendices 1, 2, 3).
The importance of TSPO for regulation of gene expression was already shown by previous studies. Microarray analysis applied in a previous study suggested regulation of cell cycle related genes by TSPO ligands [35]. For example, the TSPO ligand-mediated signaling in colorectal cancer cells involved the upregulation of the cyclin-dependent kinase inhibitors p21WAF1/CIP1 and p27Kip1, cdc16, and the cell cycle inhibitors gadd45 and gadd153, the downregulation of the cyclins D1 and B1, as well as the inactivation of ERK1/2 [35]. Other studies have shown that exogenous TSPO ligands like FGIN- 1-27, PK 11195, and Ro5-4864 dose dependently (10 μM – 100 μM) inhibited cell proliferation of human breast cancer and colorectal cancer cells measured after 72 hrs, probably via an arrest of cells in the G0/G1 phase as observed after 24 hrs, which is similar to the cell cycle effects we observed after 24 hrs [33,34]. Thus, our results are in accord with previous studies, regarding effects of particular durations of application of TSPO ligands as well as of stable TSPO knockdown.
Very interesting regarding the capability of the TSPO to regulate gene expression is the strong reduction of the expression of v-FOS, also known as c-FOS [73], that we found after TSPO knockdown and exposure to 25 μM of PK 11195 for 24 and 48 hours. Also FOSB is affected (FBJ murine osteosarcome viral oncogene homolog B; see appendices). The FOS gene family consists of 4 members: FOS, FOSB, FOSL1, and FOSL2. These genes encode leucine zipper proteins that can dimerize with proteins of the JUN family, thereby forming the transcription factor complex AP-1. As such, the FOS proteins have been implicated as regulators of gene expression for cell proliferation, differentiation, and transformation. In some cases, expression of the FOS gene has also been associated with apoptotic cell death [67]. Interestingly, the relative magnitudes of changes in v-FOS gene expression (TSPO knockdown > 48 hours of PK 11195 exposure > 24 hours of PK 11195 exposure) is in accord with our prior expectations regarding relative changes in gene expression. Here it can be also noted that from 24 hours to 48 hours of PK 11195 exposure the proportion of the group of genes related to repair and binding of DNA increases till it becomes the largest group of the total population of genes that are affected both by 48 hours of exposure to PK 11195 (25 μM) and TSPO knockdown (Figure 2). This gives further indication that regulation of DNA functions such as gene expression is one of the major TSPO functions.
In addition to v-FOS, Ran-binding protein 3 (RANBP3) gene expression was most strongly reduced by TSPO knockdown as well as by exposure to 25 μM of PK 11195 for 24 and 48 hours. RANBP3 is a ∼55-kDa protein that functions as a cofactor for Chromosome Region Maintenance (Crm1) - mediated nuclear export [46,59]. RANBP3 mediates nuclear export of Smad2 and Smad3 in a Ran-dependent manner. Depletion of RANBP3 expression enhances TGF beta-induced antiproliferative and transcriptional responses [60]. Thus, it may be that TSPO is able to regulate transport of molecules in and out of the nucleus, which may contribute to TSPO’s capability to modulate gene expression.Previously, it was suggested the transport of cholesterol by TSPO into the nucleus serves to modulate gene expression [74].
Summarizing, our data indicate firstly that TSPO is involved in regulation of expression of numerous genes related to various functions. One such function appears to be the cell cycle. Some genes and groups of genes show increasing changes in proportion to the duration of TSPO manipulation (i.e. 24 and 48 hrs of PK 11195 exposure, and permanent disabling by stable knockdown by siRNA transfection), in particular: c-FOS, RANBP3, gene expression related to repair and binding to DNA, and gene epression for small nucleolar RNAs. As the effects of TSPO on gene expression are myriad, it can be expected that more studies in this area will lead to important insights regarding tumorigenesis, and also regarding potential anti-cancer mechanisms.
We thankfully acknowledge the research support from: Niedersachsen – Israel Project (JB, WK,MG, LV). Furthermore, we want to thank Dr. Salinas-Riester and Lennart Opitz of the DNA Microarray Facility (TAL) from the University of Göttingen. Many thanks to Dr. Elisabeth Hand and Dr.Alexandra Schrader from the University Medical Center of Göttingen for support with RT-PCR experiments.
Direction, magnitude and significance of changes in expression of genes related to the cell cycle ( BRCA1 , CDC2 , CDC6, Claspin , CyclinE2 , CyclinG2 , EGR1 , KIAA , MCM2 , MCM4 ), and also of genes known to regulate gene expression and nuclear transport (FOS, RANBP3), as resulting from TSPO knockdown as well as PK 11195 exposure, as screened with microarrray.
|
|
TSPO |
PK 11195 |
PK 11195 |
knockdown |
24 hrs |
48 hrs |
||
Gene |
Target in cell cycle or nuclear function |
Microarray |
Microarray |
Microarray |
expression level |
expression level |
expression level |
||
|
(x-fold) |
(x-fold) |
(x-fold) |
|
|
|
|
|
|
BRCA1 |
S/G2 |
(+) 2.1 |
(+) 2.7 |
(+) 1.7 |
CDC2 |
G2/M |
(+) 2.5 |
(+) 1.8 |
(+) 1.7 |
CDC6 |
G1/S |
(+) 2.4 |
(+) 3.4 |
(+) 1.8 |
Claspin |
S/G2 |
(+) 2.3 |
(+) 3.1 |
(+) 1.6 |
CyclinE2 |
G1 |
(+) 2.1 |
(+) 3.5 |
(+) 1.5 |
CyclinG2 |
G2/M |
0.4 |
(-) 0.5 |
no effect |
EGR1 |
G1 |
0.4 |
no effect |
(-) 0.7 |
KIAA |
G2/M |
0.4 |
no effect |
(+) 2.1 |
MCM2 |
S |
(+) 2.3 |
(+) 2.0 |
no effect |
MCM4 |
G2M |
(+) 2.2 |
(+) 2.6 |
no effect |
v-FOS |
gene expression |
(-) 0.2 |
(-) 0.3 |
(-) 0.3 |
RANBP3 |
nuclear transport |
(-) 0.1 |
(-) 0.4 |
(-) 0.5 |
Correlations between changes in gene expression and effect on the cell cycle following TSPO knockdown by siRNA and application of PK 11195. The genes affected by our exposures to PK 11195 and by TSPO knockdown are presented in the first column. In the second, third, and fourth columns previously reported effect by the individual gene products in question on cell cycle phases is presented. In the fifth column representative references reporting the effects of the gene products in question on the cell cycle are given. The following paired columns (6 and 7; 8 and 9; 10 and 11) show the effects on gene expression and cell cycle phase by respectively, PK 11195 for 24 hours, PK 11195 for 48 hours, and TSPO knockdown by siRNA. Symbols: Black arrows pointing up mean (1) increase in numbers of cells in a particular cell phase (2) increase in gene expression. White arrows pointing up mean (1) decrease in numbers of cell in a particular cell phase (2) decrease in gene expression. Shaded cells (in pairs) show when the effects on the cell cycle phases are in accord with the effects on gene expression following exposure to PK 11195 for 24 hours, to PK 11195 for 48 hours, and due to TSPO knockdown by siRNA as can be predicted from previous reports on the effects on the cell cycle by the gene products in question.
Gene expression of non protein coding genes (for Small nucleolar RNA).
Small nucleolar RNA gene expression changes in U11MG glioblastoma cells
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
24 hrs of PK11195 |
48 hrs of PK11195 |
Permanent TSPO knockdown by stable siRNA transfection |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
|
|
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Summary:
-6-Of the 610 genes affected by 24 hours of PK 11195 exposure 6 were for small nucleolar RNAs
-12-Of the 359 genes affected by 48 hours of PK 11195 exposure 12 were for small nucleolar RNAs
-68-Of the 1249 genes affected by stable TSPO knockdown 68 were for small nucleolar RNAs
Venn diagrams of populations of significantly regulated genes as determined with microarray studies. We compared effects of TSPO knockdown (knockdown) in U118MG cells with effects of exposures for 24 hrs and 48 hrs with 25 μM of PK 11195 (in A and B, respectively) on wild type U118MG cells, using a cut off of 2-fold changes in gene expression for individual genes in each microarray assay. The observed effects of all treatments were in comparison to their respective controls i.e. Scr for TSPO knockdown and vehicle exposure for the PK 11195 exposures. We found expression of 128 genes (A) affected both by TSPO knockdown and by exposure for 24 hrs to PK 11195, and 85 genes (B) affected both by TSPO knockdown and by exposure for 48 hrs to PK 11195.
Categorization of genes according to their associated functions displaying 2-fold changes or more with TSPO knockdown as well as with PK 11195 (25μM) exposures: for 24 hrs (A) and for 48 hrs (B). Thus, these are the 128 genes and 85 genes (in A and B, respectively) as determined with TSPO knockdown and 24 hrs and 48 hours of PK 11195 exposure that were presented in Figure 1.
RT-PCR analyis of genes related to the cell cycle of U118MG TSPO knockdown cells and U118MG cells exposed to the TSPO ligand (25μM) PK 11195 for 24 hrs and 48 hrs. As a priori determined by microarray, the selected genes: 1) showed more than 2-fold differences in gene expression between TSPO knockdown cells and their Scr control cells; and also 2) showed more than 2-fold differences in gene expression between cells exposed for 24 hrs and 48hrs to 25 μM of PK 11195 and their vehicle control cells. The respective controls were set to 1 (indicated by the dotted line in the figure, with the SD values detailed in the Results’ section) and the relative expression of the cell cycle gene ofBRCA1 (A), CDC2 (B), CDC6 (C), Claspin (D), and CyclinE2 (E) is given. The changes in gene expression, in particular the directions, observed with RT-PCRbasically are in agreement with the data obtained by microarray. The y-axis indicates the relative mRNA expression level normalized to the average of the controls, with the controls are set to 1. Results arepresented as mean ± SD, n = 9,*** p < 0.001, and * = p < 0.05 versus the respective controls. The observed effects of all treatments were in comparison to their respective controls i.e. Scr for TSPO knockdown and vehicle exposure for the PK 11195 exposures.
RT-PCR analyis of genes related to the cell cycle of U118MG TSPO knockdown cells and U118MG cells exposed to the TSPO ligand PK 11195 (25μM) for 24 hrs. As a priori determined by microarray, the selected genes: 1) showed more than 2-fold differences in gene expression between TSPO knockdown cells and their Scr control cells; and also 2) showed more than 2-fold differences in gene expression between cells exposed for 24 hrs to 25 μM of PK 11195 and their vehicle control cells, and 3) did not show such effects after 48 hrs of exposure to PK 11195.
The respective controls were set to 1 (indicated by the dotted line in the figure with the SD values detailed in the Results’ section) and the relative expression of the cell cycle genes ofMCM2 (A),MCM4 (B), and CyclinG2 (C) is given. Changes in gene expression, in particular the direction, as observed with RTPCR were basically in agreement with the data obtained by microarray. As an exception, in U118MG TSPO knockdown cells, CyclinG2 (C) did not show a significant difference when analyzed with RT-PCR, but nonetheless did so after 24 hrs of PK 11195 exposure. The y-axis indicates the relative mRNA expression level normalized to the average of the controls, where the controls are set to 1. Results are presented as mean ± SD, n = 9,*** p < 0.001, ** p <0.01 versus the respective controls. The observed effects of all treatments were in comparison to their respective controls i.e. Scr for TSPO knockdown and vehicle exposure for the PK 11195 exposures.
RT-PCR analyis of genes related to the cell cycle of U118MG TSPO knockdown cells and U118MG cells exposed to TSPO ligand PK 11195 (25μM) for 48 hrs. As a priori determined by microarray, the selected genes: 1) showed more than 2-fold differences in gene expression between TSPO knockdown cells and their Scr control cells; and also 2) showed more than 2-fold differences in gene expression between cells exposed for 48 hrs to 25 μM of PK 11195 and their vehicle control cells, and 3) did not show such effects after 24 hrs of exposure to PK 11195.
The respective controls were set to 1 (indicated by the dotted line in the figure with the SD values detailed in the Results’ section) and the relative expression of the cell cycle genes ofEGR1 (A) and KIAA (B) were given. For TSPO knockdown cells, changes in gene expression, in particular the direction, as observed with RT-PCR for EGR1 (A) and KIAA (B),were basically in agreement with the data obtained by microarray. However, after PK 11195 exposure for 48 hrs U118MG cells did not show a significant difference for EGR1 expressionwhen analyzed with RT-PCR, while KIAA expression appeared increased with RT-PCR (instead of decreased as observed with microarray). The y-axes indicate the relative mRNA expression level normalized to the average of the controls, where the controls are set to 1. Results are presented as mean ± SD, n = 9,*** p < 0.001 versus the respective controls. The observed effects of all treatments were in comparison to their respective controls i.e. Scr for TSPO knockdown and vehicle exposure for thePK 11195 exposures.
RT-PCR analyis of v-FOS (A) and RANBP3(B) genes in U118MG TSPO knockdown cells and in U118MG cells exposed to TSPO ligand PK 11195 (25 μM) for 24 hrs and 48 hrs. As a priori determined by microarray, these selected genes: 1) showed a more than 10-fold difference in gene expression of RANBP3 and a more than 5-fold difference in gene expression of v-FOS between TSPO knockdown cells and their Scr control cells; 2) showed more than 2-fold changes (RANBP3) and 3-fold changes (v-FOS) in gene expression in cells exposed for 24 hrs and 48 hrs to 25 μM of PK 11195 compared to their vehicle control cells. In fact, after TSPO knockdown, v-FOS and RANBP3gene expression was affected more than of other genes,as determined with microarray (Table1, Appendices 1, 2, 3).
The respective controls were set to 1 (indicated by the dotted line in the figure with the SD values detailed in the Results’ section) and the relative RT-PCRderived observations forgene expression of v-FOS (A) and RANBP3 (B) are given. For all treatments (TSPO knockdown and PK 11195 exposure), changes in gene expression, in particular the direction, as observed with RT-PCR were basically in agreement with the data obtained by microarray. The y-axis indicates the relative mRNA expression level normalized to the average of the controls, where the controls are set to 1. Results are presented as mean ± SD, n = 9,*** p < 0.001 and ** p < 0.01 versus the respective controls.The observed effects of all treatments were in comparison to their respective controls i.e. Scr for TSPO knockdown and vehicle exposure for the PK 11195 exposures.
Quantification of cell cycle phases with the aid of PI. The relative proportion of cells in each of the G0/G1, S, and G2/M phases from U118MG Scr control cells and U118MG TSPO knockdown cells are shown in (A), WT U118MG vehicle (24 hrs exposure) control cells and cells exposed for 24 hrs to 25 μM of PK 11195 are shown in (B) and WT U118MG vehicle (48 hrs exposure) control cells and cells exposed for 48 hrs to 25 μM of PK 11195 in (C). Results are presented as mean ± SD, n = 9,*** = p < 0.001, ** = p < 0.01, and * = p < 0.05 versus the respective controls.