
Research Article
Austin J Pharmacol Ther. 2015; 3(1).1065.
Does CYP1A2 Genotype Influence Coffee Consumption?
Santos RM1*, Cotta K1, Jiang S2 and Lima DRA3
1Department of Pharmaceutical Sciences, South University School of Pharmacy, USA
2Department of Biomedical Science, Mercer University School of Medicine, USA
3Instituto of Neurology, Federal University of Rio de Janeiro, Brazil
*Corresponding author: : Santos RM, Department of Pharmaceutical Sciences, South University School of Pharmacy, 709 Mall Boulevard, Savannah, GA 31406, USA
Received: November 26, 2014; Accepted: February 05,2015; Published: February 09, 2015
Abstract
Coffee is the major source of caffeine in the American diet. Caffeine is 95% metabolized by CYP 1A2, which has a polymorphic genetic binomial distribution within the population. The homozygous wild type confers a fast metabolizer phenotype and the homozygous variant allele confers a slow metabolism of caffeine, the latter being the least predominant in the normal population. Our study objective was to examine whether genetic variability of caffeine metabolism (CYP 1A2) is associated with plasma caffeine levels and can influence the coffee consumption in young healthy volunteers. We found an inverse relationship between caffeine metabolization by CYP 1A2 and caffeine plasma levels at 45-60 minutes after intake of one cup of standardized brewed coffee (150 mg of caffeine). Our sample size was small (15 volunteers) but all possible genotypes were represented in accordance with the normal distribution in the population. We did not find a correlation between CYP 1A2 genotype and frequency or type of coffee selection (caffeinated or decaffeinated). We concluded that a bigger sample size is needed in order to identify a probable influence of caffeine metabolism phenotype on coffee consumption.
Keywords: Coffee consumption; Caffeine; Plasma levels; CYP1A2 genotype
Introduction
The association between regular coffee intake and the risk of developing a number of disorders such as myocardial infarction [1, 2], hypertension [3-6], breast cancer [7] and Parkinson’s disease [8-10], just to name a few, has been the objective of various epidemiologic studies. All of the above mentioned studies mainly focused on the content of caffeine in coffee and considered caffeine to be the main ingredient responsible for the observed effects.
Caffeine is metabolized primarily by cytochrome P450 1A2 (CYP1A2) in the liver through an initial N-demethylation [11,12]. CYP1A2 accounts for approximately 95% of caffeine’s metabolism and shows genetic polymorphism that reflects the variability in enzyme activity between individuals [13].
An A→C substitution at position 734 (CYP1A2*1F) in the CYP1A2 gene located at 15q24, decreases enzyme inducibility; as measured by the ratio of plasma or urinary caffeine to caffeine metabolites after a dose of caffeine, resulting in impaired caffeine metabolism [11,12,14]. Carriers of the variant CYP1A2*1F allele are “slow” caffeine metabolizers, whereas individuals who are homozygous for the CYP1A2*1A allele are “fast” caffeine metabolizers [11,12,14]. However, it is not clear whether CYP 1A2 genotype alters caffeine consumption and prevents slow metabolizers from drinking coffee regularly.
The fact that caffeine is one of the most widely used CNS stimulant and coffee the major source of caffeine in our diet, probably led those who are very sensitive to the effects of caffeine/coffee, to use decaffeinated coffee or to simply reduce or even abstain from coffee intake. Additionally, the levels of caffeine in the body after coffee intake are highly variable due to at least two main reasons [15-17] . The first is related to the coffee preparation itself (type/origin of the beans, method of preparation) [18]. The second one is related to individual genetic make-up in terms of caffeine pharmacokinetic metabolization (CYP1A2) and pharmacodynamic effects on adenosine receptors (A2A). Considering all the above mentioned it is tempting to hypothesize that individuals that tend to accumulate caffeine due to slow metabolization will present increased levels of caffeine in the body. These in turn will be responsible for the adverse effects that could lead those individuals to abstain or avoid using caffeinated coffee. Another possibility would be to look at the individual intervariability on adenosine receptor, more specifically receptors A2A (ADORA2) and its possible consequences on coffee effects.
Objectives
The purpose of the present study is to examine whether genetic variability of caffeine metabolism (CYP 1A2) is associated with plasma caffeine levels and can influence the coffee consumption in young healthy volunteers.
Materials and Methods
Subjects
Fifteen normal volunteers enrolled in the PharmD Program at South University School of Pharmacy, Savannah, GA. participated in this pilot research project. The study group is composed of twelve females and three males aging between 22 and 43 yrs. old and with body weights ranging from 150 to 250 lbs. The enrollment in this study was random with no specific inclusion or exclusion criteria. We conducted a brief oral presentation in class explaining the purpose of the study to the students and collected signatures of possible interested participants. The subjects enrolled in this study signed an informed consent form that was previously submitted and approved by the Institutional Review Board of South University.
Coffee preparation
One hundred grams of ground, medium roast coffee was brewed using 1.5 L of spring water under conditions set at temperature controlled brewer system BrewWISE (BUNN).
Sample collection
On the day of the study, the volunteers arrived at school having fasted for at least 8 hours. The protocol involved the collection of two time-point blood samples. The first sample was time-point zero (t0) collected as soon as they arrived. The second sample (t1) was collected between 45-60 minutes after the administration of one cup of 200 ml of a standardized brewed coffee, followed by a light breakfast composed of: a bagel with cheese spread, a muffin and a cookie. The first blood sample was collected with EDTA and used to harvest DNA for genotyping. The second blood sample was collected with heparin and used to determine the plasma caffeine levels after acute coffee intake.
DNA preparation and CYP1A2 genotyping
Two ml of whole blood collected in EDTA tubes were used for genotyping. The blood sample was centrifuged at 14,300 g for 4 min at 20°C to separate the plasma from blood cells, which were used for DNA isolation by standard procedures (ArchivePure DNA Blood Kit, cat#2300740, 5-Prime, Gaithersburg, MD). After the final step of DNA hydration, the samples were kept at 4oC for short-term storage until the determination of the genotype. The DNA purity was ascertained through nucleic acid concentration determination, utilizing the absorbance spectra of 220-320 nm and calculating the ratio of A260/ A280 as a measure of purity (NanoDrop 2000C, Thermo Scientific, Wilmington, DE).
The CYP1A2 polymorphism was detected by real-time restrictionfragment length polymorphism-polymerase chain reaction (TaqMan Drug Metabolism Genotyping Assay, Applied Biosystems, Carlsbad, CA). The assay was run in duplicates (Applied Biosystems 7900HT Fast Real-Time PCR System) at the Hoskins Cancer Center, Mercer School of Medicine laboratories in Savannah, GA. Figure 1 depicts the rationale of assigning the specific alleles for each sample according with the fluorophore that is released during the PCR reaction. In our method VIC™ dye fluoresces when Allele 1 is present, which corresponds to the variant allele CYP 450 1A2*1F; when FAM™ dye fluoresces means that Allele 2 is present and corresponds to the wild type, most common form of allele, CYP 450 1A2*1A. The phenotype will depend on the presence of a heterozygous or homozygous pair of alleles. There is co-dominance for this trait so that homozygosis for the variant will determine a slow metabolizer and homozygosis for the wild-type will determine a fast metabolizer phenotype and the presence of both alleles will determine an intermediate phenotype.
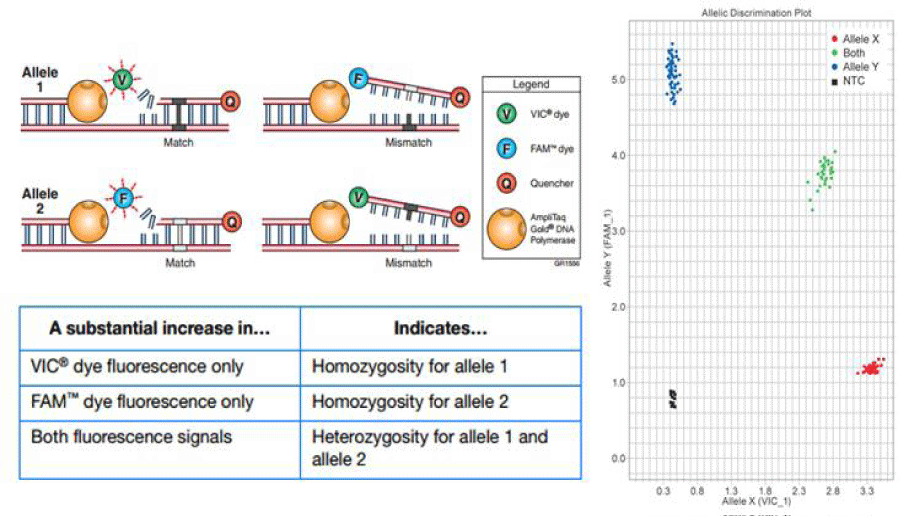
Figure 1: TaqMan Drug Metabolism Genotyping Assay. Two sets of primers with 2 different fluorophores, each one for a specific allele. Wild type represented by
the presence of fluorescent dye FAM and the variant represented by VIC fluorescence. The right panel shows the distribution of the pair of alleles, according with
the fluorescence emission region.
Plasma Caffeine Levels
Apparatus
The instrumentation utilized for chromatography included a High Performance Liquid Chromatograph Shimadzu (Model LC- 20AT, Prominence, Kyoto, Japan) equipped with a manual injection valve (Rheodyne Injector Model 7725i) and a sample loop of 100 μL (cat# 57627, SUPELCO, Sigma-Aldrich), a variable wavelength detector (Shimadzu SPD 20A), degasser and computerized remote control (Shimadzu CBM 20A/Alite) and a reverse phase column C18 particle size 5μm, internal diameter 4.6 mm, 25 cm length (Ascentis C18 HPLC column, catalog # 581325-U, Sigma-Aldrich) linked to a guard column (Ascentis C18 Supelguard cartridge, particle size 5 μm, internal diameter 3.0 mm, 2 cm length, catalog # 581375-U). Peak areas and retention time were evaluated by EZstart Software and all injections were made with a 25-μL syringe (Model 702 SNR, Hamilton Co, Reno, NV) after filtration with Millipore filter (MILLEX, Millipore Corp., Bedford, MA). The mobile phase was filtered through membrane filter (type HAWP, 0.45 μm, Millipore Corp., Bedford, MA).
Reagents
Caffeine was obtained from Sigma-Aldrich (standard grade, code C1778); Sodium Acetate Tri-hydrate (Fisher Scientific, HPLC grade); Glacial Acetic Acid (Fisher Scientific certified A.C.S.); Acetonitrile HPLC grade (Fisher Chemicals); MilliQ- Water (Millipore); Acetate Buffer pH 4.0, 10 mmol/ and Human serum (Sigma-Aldrich).
Standard curve and samples preparation
A standard stock solution was made by weighing 10 mg of caffeine and dissolving it in 100 ml of acetate buffer 10mmol/L, pH 4.0. The primary standard concentration was of 100 μg/mL and then diluted with human serum to obtain an intermediate standard solution of 20 μg/mL. An 8-time point standard curve was prepared by sequential dilution of the intermediate standard with human serum, ranging from 10 to 0.078 μg/mL. A blank was prepared with human serum and distilled water. All the standards were carefully vortexed for 10” and centrifuged at 12, 800 g per 10 minutes and the supernatant injected in the HLPC apparatus after filtration through membrane filters (SLGV033RB, MILLEX 0.22 μm). Two hundred and fifty microliters of plasma obtained from each of the blood samples of the 15 healthy volunteers were brought to room temperature, vortexed for 10” and centrifuged at 12,800 g for 10 minutes. The supernatant was then filtered and 10 μL of the filtrate was injected into the HPLC apparatus.
HLPC method
The method used was based on Blanchard J and col. [19]. Briefly, standards and samples were scanned over 10 minutes at 273 nm wavelength. The mobile phase constituted of a 10mM acetic acid/ acetate buffer, pH 4.0, 85/15, v/v, buffer/acetonitrile. An isocratic HPLC run with a flow of 1.0 mL/min and a background pressure of approximately 2000 psi.
Standards in duplicate were injected for each of the 8-time points. The standard curve constructed with the average value produced a very accurate linear regression line and a slope with a r2 = 0.9902 (CV% 0.023 and SD% 3.58).
Results
Plasma caffeine levels
The mean plasma caffeine level (n=15) was 0.4 mg/L, the levels ranging from zero or below the minimum limit of detection (LOD= 0.01 mg/L) to 1.3 mg/L. The plasma caffeine levels found in this pilot study are in accordance with previous studies. These studies indicated a range between 0.25-2.0 mg/L [20], 0.5-10.0 mg/L [21], 0.28-10.58 mg/L [22], 0.05-4.94 mg/L [23] of plasma caffeine levels after coffee (1-2 cups) or caffeine (200-350 mg) intake. A typical plasma chromatogram is shown on Figure 2. Peaks are numbered from 1-4, caffeine represented by peak 4 with a retention time of 5.5 minutes on average. The other 3 peaks, even though not standardized for the purpose of our study, are likely to represent the 3 main metabolites of caffeine: theophylline (4%), theobromine (12%) and paraxanthine (84%) [17]. Under the chromatographic conditions used in our study, they are eluted in the following sequence: theobromine, paraxanthine and theophylline (peaks 1-3, respectively) [19].
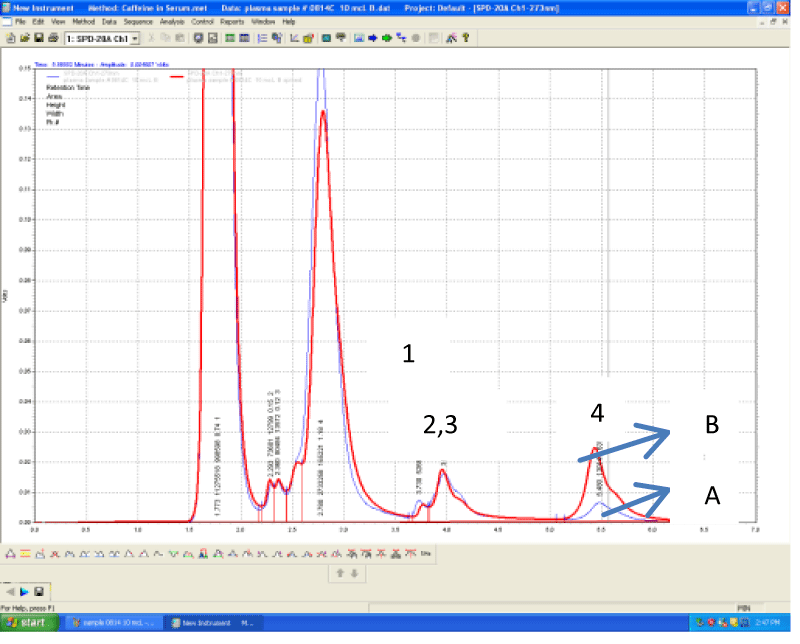
Figure 2: An overlay of chromatograms of plasma sample from a healthy volunteer 45 minutes after drinking one cup of standardized brewed coffee (A) and the
same sample spiked with 10 μL of standard solution of caffeine of 1 mg/L (B). Peak 1= theobromine, Peak 2= paraxanthine, Peak 3 (shoulder) = theophylline, Peak
4= caffeine. Retention times are 3.7, 3.9, 4.2 and 5.5 respectively.
Caffeine metabolism and CYP 1A2 genotyping
Eleven out of fifteen blood samples produced a satisfactory DNA quality to perform the genotyping assay. We found that 8 out of 11 (73%) presented a homozygous genotype for the wild type allele (1A2 *1A), which translates into the fast metabolizer phenotype, the most common in the normal population [17]. Only 1 out of 11 (9%) presented a homozygous genotype for the variant type allele (1A2 *1F), slow metabolizer phenotype. There were 2 out of 11 (18%) heterozygous genotypes that can be translated into intermediate metabolizers (Table 1).
Sample ID
Caffeine Plasma levels (mg/L)
CYP 1A2
Genotype
Metabolizer
Phenotype
Coffee
Drinker
Cups of coffee/day
0214C
0.39
*1F/*1A
Intermediate
yes
1 cup/day
0314C
0.01
*1A/*1A
Fast
no
none
0414C
0.67
*1A/*1A
Fast
no
none
0514C
0.16
*1A/*1A
Fast
yes
1 cup every other day
0614C
0.16
*1A/*1A
Fast
no
none
0714C
0.11
*1F/*1A
Intermediate
yes
1 cup/week
1114C
1.11
*1F/*1F
Slow
yes
1 cup/day
0914C
0.23
*1A/*1A
Fast
yes
3 cups/week
1014C
0
*1A/*1A
Fast
no
none
0814C
0.44
*1A/*1A
Fast
yes
1 cup/day
1214C
0.48
*1A/*1A
Fast
yes
2 cups/day
*Allele 1 = CYP 450 1A2*1F (variant) Allele 2= CYP 450 1A2*1A (wild type)
Table 1: The genotype and corresponding phenotype (slow, intermediate and fast metabolizer) are shown and the plasma levels of caffeine obtained after an acute dose of caffeine (approx. 150 mg) administered as a cup of standardized brewed coffee. Answers to brief questionnaire on coffee consumption are also shown. Note: Only 11 out of 15 blood samples produced a satisfactory DNA quality to perform the genotyping assay.
Habitual Coffee Consumption
The volunteers filled a small questionnaire during the breakfast prior to drawing the second blood sample. The questionnaire consisted of a few questions about the number of cups of coffee, and coffee drinking frequency; the type of coffee used, if caffeinated or decaffeinated and the current use of any regular medication prescribed or over the counter. The results related to habitual coffee consumption are summarized on Table 1. Fifty percent of the fast metabolizers were not coffee drinkers and the only slow metabolizer consumed 1 cup daily.
Discussion
It is a common belief that people avoid drinking coffee because of its effects, even though they also refer to the aroma of coffee as something very pleasant [22]. The caffeine content in coffee is likely to be responsible for the side effects experienced, since the common complaints are hand shaking, increase in the heart rate and anxiety [15,24]. Such effects are typically related to central nervous stimulation, one of the important properties of caffeine [20].
Our study is the first, from our knowledge, to confirm that individuals that have the polymorphic variant genotype (1A2 *1F) for caffeine metabolism (CYP 1A2), which translate into slow metabolizer phenotype, displayed the highest plasma caffeine level after administration of one cup of coffee. The results also indicated that 8 out of 11 (90%) of the volunteers had the wild type allele (fast metabolizer), which is the most commonly found in the normal population [15]. It is interesting to notice that the fast metabolizers showed great variability in their plasma caffeine levels, which ranged from zero (not detectable) to 0.67 mg/L. The two intermediate metabolizers had plasma caffeine levels of 0.11 and 0.39 mg/L and the slow metabolizer had a plasma caffeine level of 1.11 mg/L (see Table 1).
One of the limitations of this study was that as a pilot study we established a minimum of twelve healthy volunteers with 10-20 % more to account for any losses during the study (N=12 + 3). The rationale for selection of 15 students as sample size is based on the average number of healthy volunteers normally enrolled on phase I clinical trials [25]. For future studies, we intend to make the research method less invasive, more attractive to students to participate, and thus increase the sample size. In order to do that we decided to use saliva instead of blood to measure caffeine levels and buccal cotton swab as a way to obtain cells to harvest DNA for genotyping.
We could not find a direct relationship between the presence of the CYP 1A2 variant allele and reduced intake of coffee/caffeine. A previous study [15] examined the relationship between slow caffeine metabolizer and habitual caffeine consumption and did not find any association. Instead, they found a positive correlation with the polymorphic receptor of adenosine ADORA 2A, main target of caffeine action in the central nervous system. More recently, genomewide association analysis (GWAS) of coffee drinking, where coffee consumption was used as a model for addictive behavior, analyzed 8 Caucasian cohorts (N= 18,176). Their data pointed to regions on chromosome 15 (15q24) of other single-nucleotide polymorphisms (SNPs) between the genes CYP1A1 and CYP1A2, but no association was found between variants of CYP1A2 [26]. At the turnaround of year 2014, a consortium was formed, named The Coffee and Caffeine Genetics Consortium, looking for genome-wide identification of possible loci associated with coffee consumption. They found within a population of European and African-American ancestry that six loci showed positive association with coffee consumption and two of them, AHR and CYP 1A2 are directly related with caffeine metabolism [27].
Conclusion
Our results did indicate an inverse relationship between caffeine metabolism (CYP 1A2) and plasma caffeine levels. However due to our small sample we were unable to detect a correlation between the genetic variability of caffeine metabolism (CYP 1A2) and the coffee consumption habit in young healthy volunteers. Nevertheless, the GWAS have the advantage of surveying a very large population allowing them to attribute relevance to a potential influence of genetics on coffee consumption [26,27]. Our hypothesis is in conformity with their findings but further studies with a larger population are needed to better understand the correlation between CYP1A2 genotype and coffee consumption.
Acknowledgement
We appreciated the donation of the BrewWise brewer from BUNN, and we acknowledge the support of our librarian Valerie Yaughn, and her assistant Miriam Wing, whom were real partners providing us promptly with all the articles necessary to put the manuscript together.
References
- Mukamal KJ, Hallqvist J, Hammar N, Ljung R, Gémes K, Ahlbom A, et al. Coffee consumption and mortality after acute myocardial infarction: the Stockholm Heart Epidemiology Program. Am Heart J. 2009; 157: 495-501.
- Richardson T, Baker J, Thomas PW, Meckes C, Rozkovec A, Kerr D. Randomized control trial investigating the influence of coffee on heart rate variability in patients with ST-segment elevation myocardial infarction. QJM: monthly journal of the Association of Physicians. 2009; 102: 555-561.
- Geleijnse JM. Habitual coffee consumption and blood pressure: an epidemiological perspective. Vasc Health Risk Manag. 2008; 4: 963-970.
- Palatini P, Ceolotto G, Ragazzo F, Dorigatti F, Saladini F, Papparella I, et al. CYP1A2 genotype modifies the association between coffee intake and the risk of hypertension. J Hypertens. 2009; 27: 1594-1601.
- Zhang Z, Hu G, Caballero B, Appel L, Chen L. Habitual coffee consumption and risk of hypertension: a systematic review and meta-analysis of prospective observational studies. Am J Clin Nutr. 2011; 93: 1212-1219.
- Zhao Y, Wang J, Ballevre O, Luo H, Zhang W. Antihypertensive effects and mechanisms of chlorogenic acids. Hypertens Res. 2012; 35: 370-374.
- Kotsopoulos J, Ghadirian P, El-Sohemy A, Lynch HT, Snyder C, Daly M, et al. The CYP1A2 genotype modifies the association between coffee consumption and breast cancer risk among BRCA1 mutation carriers. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007; 16: 912-916.
- Ragonese P, Salemi G, Morgante L, Aridon P, Epifanio A, Buffa D, et al. A case-control study on cigarette, alcohol, and coffee consumption preceding Parkinson's disease. Neuroepidemiology. 2003; 22: 297-304.
- Tan EK, Chua E, Fook-Chong SM, Teo YY, Yuen Y, Tan L, et al. Association between caffeine intake and risk of Parkinson's disease among fast and slow metabolizers. Pharmacogenet Genomics. 2007; 17: 1001-1005.
- Popat RA, Van Den Eeden SK, Tanner CM, Kamel F, Umbach DM, Marder K, et al. Coffee, ADORA2A, and CYP1A2: the caffeine connection in Parkinson's disease. Eur J Neurol. 2011; 18: 756-765.
- Butler MA, Iwasaki M, Guengerich FP, Kadlubar FF. Human cytochrome P-450PA (P-450IA2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proceedings of the National Academy of Sciences of the United States of America. 1989; 86: 7696-7700.
- Gu L, Gonzalez FJ, Kalow W, Tang BK. Biotransformation of caffeine, paraxanthine, theobromine and theophylline by cDNA-expressed human CYP1A2 and CYP2E1. Pharmacogenetics. 1992; 2: 73-77.
- Sachse C, Brockmöller J, Bauer S, Roots I. Functional significance of a C-->A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br J Clin Pharmacol. 1999; 47: 445-449.
- Han XM, Ou-Yang DS, Lu PX, Jiang CH, Shu Y, Chen XP, et al. Plasma caffeine metabolite ratio (17X/137X) in vivo associated with G-2964A and C734A polymorphisms of human CYP1A2. Pharmacogenetics. 2001; 11: 429-435.
- Cornelis MC, El-Sohemy A, Campos H. Genetic polymorphism of the adenosine A2A receptor is associated with habitual caffeine consumption. Am J Clin Nutr. 2007; 86: 240-244.
- Djordjevic N, Ghotbi R, Jankovic S, Aklillu E. Induction of CYP1A2 by heavy coffee consumption is associated with the CYP1A2 -163C>A polymorphism. Eur J Clin Pharmacol. 2010; 66: 697-703.
- Yang A, Palmer AA, de Wit H. Genetics of caffeine consumption and responses to caffeine. Psychopharmacology (Berl). 2010; 211: 245-257.
- Mandel HG. Update on caffeine consumption, disposition and action. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2002; 40: 1231-1234.
- Blanchard J, Mohammadi JD, Conrad KA. Improved liquid-chromatographic determination of caffeine in plasma. Clin Chem. 1980; 26: 1351-1354.
- Fredholm BB, Bättig K, Holmén J, Nehlig A, Zvartau EE. Actions of caffeine in the brain with special reference to factors that contribute to its widespread use. Pharmacol Rev. 1999; 51: 83-133.
- Zylber-Katz E, Granit L, Levy M. Relationship between caffeine concentrations in plasma and saliva. Clin Pharmacol Ther. 1984; 36: 133-137.
- Alvi SN, Hammami MM. Validated HPLC method for determination of caffeine level in human plasma using synthetic plasma: application to bioavailability studies. Journal of chromatographic science. 2011; 49: 292-296.
- Holland DT, Godfredsen KA, Page T, Connor JD. Simple high-performance liquid chromatography method for the simultaneous determination of serum caffeine and paraxanthine following rapid sample preparation. Journal of chromatography B, Biomedical sciences and applications. 1998; 707: 105-110.
- Cornelis MC, El-Sohemy A, Campos H. Genetic polymorphism of CYP1A2 increases the risk of myocardial infarction. J Med Genet. 2004; 41: 758-762.
- Julious SA. Sample size of 12 per group rule of thumb for a pilot study. Pharmaceut Statist. 2005; 4: 287-291.
- Amin N, Byrne E, Johnson J, Chenevix-Trench G, Walter S, Nolte IM, et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol Psychiatry. 2012; 17: 1116-1129.
- The Coffee and Caffeine Genetics Consortium, Cornelis MC, Byrne EM, Esko T, Nalls MA, Ganna A, Paynter N, et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry. 2014.