
Research Article
Austin J Pharmacol Ther. 2021; 9(2).1134.
Genome Wide Characterization & Phylogenetic Analysis of TNF Genes in Homo Sapiens
Saif S¹, Abaidullah M²*, Mazhar MW¹, Ijaz A³, Sikandar M¹, Waqas N¹, Mahmood J¹, Aslam H¹
¹Department of Bioinformatics and Biotechnology, Government Collage University, 38000 Faisalabad, Pakistan
²Natural Medicine Research Center, College of Veterinary Medicine, Sichuan Agricultural University, China
³National Center of Excellence in Molecular biology, University of Punjab Lahore, Pakistan
*Corresponding author: Muhammad Abiadullah, Natural Medicine Research Center, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu 611130, China
Received: April 17, 2021; Accepted: May 06, 2021; Published: May 13, 2021
Abstract
Tumor Necrosis Factor is very important inflammatory signaling unit that do an important role in immune system. It functions by attachment and stimulation of different receptor’s Cysteine Rich Domains (CDRs). A number of TNF receptors mediated factors have been identified having a major role in signal transduction pathways of TNF gene family. There are about 18 TNF homologues that are identified as a major cause of many disorders like cancer, Diabetes, AIDS and many other lethal inflammations. In this study the genome wide identification of TNF gene was done. Different tools and databases were used. Identification of conserved domains was done by using pfam and homology analysis showed that the TNF might be a member of TRAFs superfamily. Structural analysis of gene showed the number of introns and exons by a three-dimensional structure of TNF gene. The TNF gene family's exon-intron structure was found to be very similar in this study. The distribution of genes across chromosomes, on the other hand, was extremely varied. Collectively, the newly discovered genes can provide a wealth of information for manipulating the TNF genome to develop drugs and strategies to treat a variety of diseases.
Keywords: Tumor necrosis factor; TNF gene family; TNF receptors; Transcription factor; Gene structure; Immune system
Introduction
TNF is a signaling molecule that is involved in a number of cellular operations, like cell division, characterization, and apoptosis having critical a role in a number of diseases. The molecular mechanisms of TNF action have been studied widely in current years. Many lab studies have shown that TNF mediates a wide range of biological operations. TNF associated different biological responses are performed by generating number of signaling pathways, as a result of different laboratory studies. NF-B, a transcription factor and AP-1, in particular, play an important function in communicating these cellular responses [1,2]. About two decades ago it was observed that TNF is a protein that is promote by the immune system can suppress the Tumor Cell proliferation. Many comprehensive studies have revealed that TNF is a significant intermediary of infections, replication of viruses, spread of tumors, rejection of transplant, many different septic shocks and Rheumatoid Arthritis, Children’s Pertussis, Down’s Syndrome etc. as shown in Figure 1. Depending upon type of target cells tumor necrosis factor can cause destructive & apoptotic death of cell. Necrosis may result in swelling of the cells, death and breakage of different organelles of cells. While as a result of apoptosis, the cell loses its shape, development of apoptotic fragments, and particular internucleosomal fragmentation of DNA occurs. It is still doubtful whether DNA fragmentation causes or results from TNF-associated cell death [3,4].

Figure 1: Major Activities Refrerd to the TNF super family.
It has been reported that there are 18 TNF homologues with 15- 20 % identity to each other since its discovery, that are responsible for a number of disorders counting arthritis Cancer, Allergy, AIDS, Diabetes and Atherosclerosis, graft versus host disease. TNF cytokines shows their expression by [4]. The activating NF-B transcription factor, c-Jun N-terminal kinase, cellular division and apoptosis [5]. As a consequence, agents that can either inhibit or block the action of these cytokines have a curative potential for diverse number of inflammations. TNF ligands have significance in the structure and functioning of immune responses, and have been linked to the development of a number of acquired and genetic diseases as shown in Table 1 [6].
Shocks
T Cells associated fatal shocks
Candida septic shock syndromeAutoimmune
Rheumatoid Arthritis
Crohan’s disease
Multiple SclerosisParasitic disease
Plasmodium Vivax Malaria
Dysentery
Children’s PertussisBacterial Disorders
Bacterial Meningitis
Mycobacterium Tuberculosis
Pre-term labor induced by InfectionMany others
Down’s Syndrome
Diabetes
Hairy cell Leukemia
Gynecological malignancy
Table 1: Some major Disorders Associated with TNF [13].
Ligands of Tumor necrosis factor partake a structural motif known as the homology domain of TNF that attaches to receptor’s (CRDs) [6]. The heterogeneity of TNF gene family is due to the CRDs that are made up of structural components, which vary in number and form. Some protein families have folds similar to the THD and CRD, increasing the likelihood that the path of interaction of gene TNF and its receptors is maintained in many other factors [6]. Presence of Cysteine-Rich Domains (CRDs) is important for the differentiation of TNF Domains which are pseudo-repeats consist of 6 residues of cysteine residues that are joined in the form of three IMG-20210316-WA0073 disulfide bonds. Amount of CRDs in a given receptor differ from 1 to 4, with the exception of CD30, which has three CRDs that have been slightly duplicated in human’s sequence but not in the sequence of mouse. A slightly rounded ladder of disulfides bridges maintains the enlarged form of TNF Receptors.
TNF dysregulation, on the other hand, has been related to a broad range of diseases). As a result, TNF is an excellent target for drug growth. The signal transduction pathways involved in transduction of signals for tumor necrosis factor gene family and the results of deletion of the genes involved in these pathways used by members of the TNF family have been studied. Our existing knowledge for signal transduction mechanisms for TNF and other different members of this family can be used to improve therapeutics. It's less clear how to build therapeutics based on other TNF superfamily members at this time.
Materials and Methods
Sequence retrieval of TNF GENE
Retrieval of DNA and protein sequence of the gene TNF in Homo sapiens from all NCBI databases, FASTA format of the TNF protein and BLAST-P was run for Homo sapiens genome database to find similar sequences from protein databases. In order to perform this protein sequence (NP_000585.2) of Tumor necrosis factor [Homo sapiens (Humans)] was given as query sequence. Target sequences then used in further databases like PFAM databases to find out the conserved domains and motifs.
Identification of domains and motifs
The sequence of TNF protein was given as a query sequence and downloaded all target sequences in order to analyze the conserved motifs and domains of the protein sequences among (Homo sapiens), (Pan troglodytes), (Rhinophitheus roxellana) and (Trachypithecus fancoisi) at NCBI online database Pfam. (https://pfam.xfam.org) and SMART (smart.embl-heidelberg.de) [7]. This step was done to remove sequences in which there were no conserved domains and motifs that are essential for TNF protein to perform its functions normally [1]. Only one conserved domain was identified in this Homo sapiens, pfam00229 that is a member of the superfamily cl00147. All of the variants were carefully examined with E-value less than 1e-10 and those with lengthy ORF were chosen for further data analysis using various databases, (http://www.ncbi.nlm.nih.gov/Structure/ cdd/wrpsb.cgi), (http://smart.embl-heidelberg.de/) and (http://pfam. janelia.org/). Position of chromosome, length of protein and cDNA and genomic sequences were observed by using NCBI database.
Gene structure and phylogenetic analysis of TNF gene family
cDNA and sequence of genome were taken from NCBI and TNF gene structure was observed by using an online tool named as Gene structure display sever(http://gsds.cbi.pku.edu.cn/) to determine the position of introns and exons and three dimensional structure of protein was obtained in TNF gene [8].
By using the bioinformatics tool MEGA7 with default parameters all the sequences from different organism like, (Homosapiens), (Pan troglodytes), (Pango abelii), (Hylobates moloch), (Macaca Mulatta), (TNF-alpha (Homo sapiens) and (Nomascus leucogenys) were aligned to perform multiple alignment and pairwise alignment in order to report the conserved domains in TNF gene members. MEGA7 was used to coordinate sequences and a perfect unrooted evolutionary tree was built on the bases of alignments of sequences. Even after that, the TNF gene family was classified based on their evolutionary similarity [9]. A separate instrument was used to measure the molecular weight and isoelectric point toolbox (http://expasy.org/tools/). Evolutionary analysis was used to name the subfamilies of tumor necrosis factor gene based on their homologs. The evolutionary relationship among the families was performed using the online Time Tree Server [10,11].
Identification of restriction sites
Genomic sequence of the TNF gene was given as a query sequence and computational analysis of the gene sequence was done by using a software, serial cloner. Number of restriction sites for different restriction enzymes were identified by developing the graphical map of given nucleotide sequence.
Analysis and Results
There were 18 different TNF genes identified in Homo sapiens by using different tools and databases all the information was presented by a supplementary data table, such as RNA, DNA and proteins sequences. The position of gene was found on chromosome number 6. There are 4 exons and only 1 conserved domain was identified. There were 4 significant A matches and 28 were insignificant A matches as a result of pfam, as shown in Figure 2. Evolutionary relationship shows the similarity of different genes, as given in Table 2.S1 [8,12].
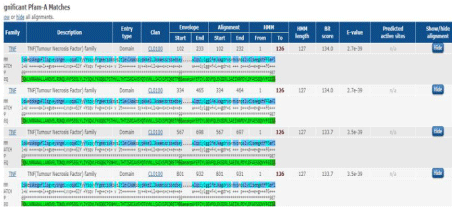
Figure 2: Significant pfam results.
Homology analysis and phylogenetic analysis: For determination of conserved domains BLAST was performed with the sequence of TNF homologues. As a result of BLAST there was one conserved domain pfam00229 ranging from 103-233 amino acid sequence, which is the most conserved domain among TNF protein members [1].
Pfam results
Trachypithecus fancoisi 033072455.1, 033216564.1
4 Un (unplaced Scaffold)
705 234 31615357
Pfam00229
Homology and phylogenetic analysis
After that comparison of sequences of TNF protein sequence was done with the identical sequences of different species (Homo sapiens), (Pan troglodytes), (Rhinophitheus roxellana) and (Trachypithecus fancoisi) at NCBI. The result of this analysis revealed that there was not high level of resemblance of TNF protein with the other, but there was but pfam00229 was highly conserved in all the species. In order to determine the evolutionary relationship of TNF (homo sapiens with other corresponding species a unrooted tree was constructed based on neighbor-joining procedure using bootstrap of 1000 replicates of 7 members, (Homosapiens), (Pan troglodytes), (Pango abelii), (Hylobates moloch), (Macaca Mulatta), TNF-alpha (Homo sapiens) and (Nomascus leucogenys) of TNF family among different species, as shown in Figure 3 [1].
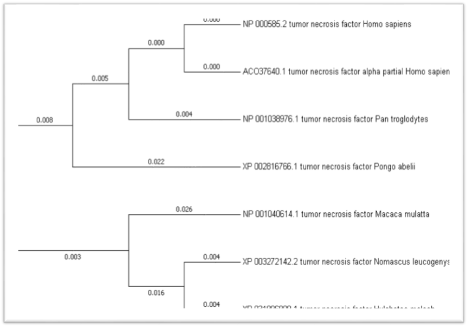
Figure 3: Phylogenetic analysis by MEGA7 software.
Gene structure analysis
(Figure 4).

Figure 4: Analysis of Gene structure.
Discussion
There are many different signaling molecules that are engaged with immune systems like tumor necrosis factor molecules. There are other different signaling molecules and pathways having a role immune regulation, like TNF molecules specifically on the downstream factor TRAF. The TRAF superfamily is important in the signal transduction pathways of TNF. Recent investigations have revealed it that the TRAF2 have very important role in both immune systems like innate and adaptive. For a number of disorders. Some other member of this group, RANKL, regulates bone resorption and can therefore be used as a pharmaceutical research target. Since RANKL transmits its signals by using, receptors such as TRAF6 and RANK could also be used to transmit its signals. TRAF6 could also be used as drug targets in the development of anti-osteoporosis drugs. At this time, it's unclear which members of the TNF superfamily are involved. to develop the therapeutic agents based on the findings of others? [13]. Under this article, we focused on those physiological pathways that might be involved in the process of TNF-mediated cytotoxic effects. And describe the situation TNF immunogenicity in human cells may be triggered by biochemical processes. TNF activates competitive pathways as it binds to target cells: encoding of defensive genes vs. a self-destruction method. Protein kinases and phospholipases are activated via receptor cluster formation, likely through optimization of a pertussis toxin-sensitive G protein. Triacylglycerol, phosphatidic acid, inositol phosphates, Arachidonic acid, triacylglycerol and other compounds are formed as a result. As a result, the most important question is: how is selectivity achieved? In addition, several other crucial questions about TNF cytotoxicity remain unanswered. What function do the various TNF receptors play, for example? Is phosphorylation of receptors important? Is there a connection between TNF receptors and signaling molecules? Further researches will hopefully develop a comprehensive sketch of the intracellular signal transduction system that can links TNF cell surface receptors to TNF cytotoxicity for tumor cells [3].
Conclusions
TNF was originally known as Tumor Suppressor, but the current studies have revealed that it is involved in diverse responses. Most important function performed by TNF gene is to control the immune system by providing immunity against bacteria & viruses. dysregulation of TNF gene, on the other hands, has been linked to a wide range of inflammations. As a result, TNF is an ideal victim for the processing and development of drugs against different diseases. Antibodies against the TNF gene and its receptors have been approved for controlling many diseases like rheumatoid arthritis and Crohn’s disease.
The exon-intron structure of the TNF gene family was found to be very similar in this study. However, there was a wide range of gene distribution across chromosomes. The discovered genes could provide a wealth of knowledge for manipulating the TNF genome in order to develop drug.
References
- Hu W, et al. "Identification and characterization of a TNF receptor-associated factor in Dugesia japonica." Gene. 2019; 681: 52-61.
- Liu Z-g. "Molecular mechanism of TNF signaling and beyond." Cell Research. 2005; 15: 24-27.
- Beyaert R, Fiers W. "Molecular mechanisms of tumor necrosis factor-induced cytotoxicity: what we do understand and what we do not." FEBS letters. 1994; 340: 9-16.
- Jarosz-Griffiths HH, et al. "TNF receptor signalling in autoinflammatory diseases." International immunology. 2019; 31: 639-648.
- Suganuma M, et al. "Carcinogenic role of tumor necrosis factor-a inducing protein of Helicobacter pylori in human stomach." BMB Reports. 2006; 39: 1-8.
- Bodmer J-L, et al. "The molecular architecture of the TNF superfamily." Trends in biochemical sciences. 2002; 27: 19-26.
- Liu T, et al. "Genome-wide characterization of TNF receptor-associated factors in the Chinese soft-shelled turtle Pelodiscus sinensis and their expression profiling in response to Aeromonas hydrophila challenge." Fish & shellfish immunology. 2020; 101: 88-98.
- Ahmad B, et al. "Genome-wide identification and expression analysis of two component system genes in Cicer arietinum." Genomics. 2020; 112: 1371- 1383.
- Biswas G, et al. "Evolutionary evidence of tumor necrosis factor super family members in the Japanese pufferfish (Takifugu rubripes): comprehensive genomic identification and expression analysis." Marine genomics. 2015; 22: 25-36.
- Li K, et al. "The involvement of TNF-a and TNF-β as proinflammatory cytokines in lymphocyte-mediated adaptive immunity of Nile tilapia by initiating apoptosis." Developmental & Comparative Immunology. 2021; 115: 103884.
- Hu W, et al. "Genome-wide identification and expression analysis of the NAC transcription factor family in cassava." PLoS One. 2015; 10: e0136993.
- Savan R, et al. "A novel Tumor Necrosis Factor (TNF) gene present in tandem with the TNF-a gene on the same chromosome in teleosts." Immunogenetics. 2005; 57: 140-150.
- Aggarwal BB, et al. "The role of TNF and its family members in inflammation and cancer: lessons from gene deletion." Current Drug Targets-Inflammation & Allergy. 2002; 1: 327-341.