
Review Article
Austin J Pharmacol Ther. 2021; 9(4).1144.
Genome Wise Identification and Phylogenetic Analyses of NOTCH1 Gene
Mazhar MW*, Sikandar M, Saif S, Mahmood J, Aslam H, Mazhar F, Tahir H, Raza A and Shehzad MB
Department of Biotechnology and Bioinformatics, Government College University, Faisalabad, Pakistan
*Corresponding author: Muhammad Waqar Mazhar, Department of Biotechnology and Bioinformatics, Government College University, Faisalabad, Pakistan
Received: June 05, 2021; Accepted: June 28, 2021; Published: July 05, 2021
Abstract
The NOTCH gene encode transmembrane receptor. It play a vital role in several process stem cell maintenance and differentiation during embryonic and adult development. When ligand bind at a specific part intracellular part of NOTCH receptor is cleaved and translocate to the nucleus from where it can bind to transcription site. NOTCH activity can promotes tissue growth and cancer in some conditions but they also suppress tumors formation in others. Their gene structure show the amount of introns and exons by a dimensions structure of NOTCH gene. (Powell, Passmore et al. 1998)Various tools or database such as Mega7, Pfam and Gene structure and display server are used to analyze their phylogeny and their chromosome positions gene structure and introns and exons. Further studies are made to target the NOTCH pathway on growth and cancer suppressor.
Keywords: Oncogenes; NOTCH1-4 receptors; Heterodimeric protein
Introduction
NOTCH gene is evolutionary conserved signaling gene that is essential for embryonic development in all metazoan organisms. It is also present in human in which it encode single pass transmembrane receptor. Many lab studies have shown that NOTCH protein mediates a wide range of biological operations. NOTCH1 gene make a protein called NOTCH1 (Borggrefe and Oswald 2009). Receptor protein have such site in which certain other proteins called ligands can attached. Attachment of the ligands to NOTCH1 can send signal throughout the body for normal development of many tissues before and after birth. It can turn normal cell into cancerous and also considered as oncogenes. It involves in cell proliferation, cell differentiation, maturation and apoptosis and cell growth.
Proposed name
Gene locus
Protein accession no.
RNA accession no.
Exons
Chromosome no.
ORF length
Amino acid length
Start of genomic location
Conserved domain in protein sequence
Pan troglodytes
CK820_G0012017
XP_009455997.1
XM_009457722.3
34
9
7440
2479
10872668
Smart000034
Pfam00008
Pfam00066
Pfam07684
Pfam12796
Pfam11235
Pfam08168
Pongo abelii
CR201_G0049823
XP_024108478.1
XM_024252710.1
34
9
7671
2556
110450912
Cd0054
Pfam00066
Pfam00088
Sd00045
Pfam11936
Pfam12796Macaca mulatta
Unknown
XP_014971839.1
XM_015116353.2
34
15
7671
2556
1699397
Cd00054
Cd00204
Pfam0066
Pfam07684
Pfam12796
Pfam06816Macaca nemestrina
unknown
XP_011723094.1
XM_011724792.2
34
Unknown
7671
2556
9666387
Cd00054
Cd00204
Pfam00066
Pfam06186
Pfam12796
Pfam07684
Sd00054
Table 1: Important feature of NOTCH1 gene.
NOTCH family encodes single pass transmembrane proteins. In mammals four NOTCH receptors are present NOTCH1, NOTCH2, NOTCH3, and NOTCH4 (Fleming 1998). These are heterodimeric protein consist of N terminal extracellular portion which are noncovalently bound to transmembrane domains. These extracellular domains of protein family consist of large numbers of tandemly repeated copies of an epidermal growth factor. NOTCH receptors interact with membrane bound ligand that are that are encoded by Jagged (JAG1 and JAG2) and Delta gene family. The signal induced by ligand binding is transmitted intracellularly. The receptor/ligand interaction induces two additional proteolytic cleavages that free the intracellular domain of the Notch receptor from the cell membrane. The cleaved fragment translocate to the nucleus due to the presence of nuclear localization signals. Once in the nucleus, the Notch intracellular domain forms a complex with the RBPSUH protein, a sequence-specific DNA binding protein (also known in mammals as CSL, CBF1 and RBP-J).
It involves in various kind of diseases such as cancer, critical congenital heart disease, carcinoma, Alagille syndrome and various others. NOTCH gene have other name AOS5, TAN1, HOVD1, HN1, Neurogenic locus Notch homolog protein. NOTCH involved in controlling of various developmental process by controlling cell divisions. Its ability to affect the cell cycle kinetics and response to apoptotic signals that notch proteins involved in the malignant transformation of some cells. In the last few years evidence has collected on notch participation in carcinogenesis and human tumors. Notch signaling is constitutively activated in several types of cancer cells and it is regarded as an ant apoptotic and pro-oncogenic signal. Notch3 overexpression are responsible for increased in vitro tumor cell growth in human lung cancer. Moreover, increased expression of Notch3 has been observed in spontaneous human pancreatic tumors. Notch signaling also play role in the induction of terminal differentiation and growth arrest.
NOTCH gene involved in broad range of disease. NOTCH pathway can cause various inherited disease. Mouse models have made for each type of disease and analyze the model lead to several potential sights. Further studies in NOTCH pathway mutant mice will lead to additional way to the pathogenesis of the disease in humans.
NOTCH receptors
NOTCH1: It is a human gene encoding single transmembrane receptors. When NOTCH1 activate before birth it induce radial glia differentiation. Reelin and NOTCH1 cooperate in the development of dentate gyrus.
NOTCH2: NOTCH2 is a protein that is encoded by NOTCH2 gene. It is associated with Alagile syndrome and Hajdu-Cheney syndrome. It can remove PEST domain and remove nonsense mediated mRNA.
NOTCH3: It encodes the third discovered human homologue of the Drosophila melanogaster type I membrane protein notch. In Drosophila, notch interaction with its cell-bound ligands (delta, serrate) establishes an intercellular signaling pathway that plays role in neural development.
NOTCH4: It is located on 6 chromosome. Its biological significance has not been described. Mutation in NOTCH4 gene associated with susceptibility to schizophrenia.
Materials and Methods
For retrieval of protein and DNA NOTCH gene from all database of the NCBI and then FASTA sequence was downloaded by running BLAST. To find similar sequence from protein database BLAST-P was run. Protein sequence was used as a query sequence. Genomic information related to gene was taken from NCBI. Targeted sequence are further used in different databases to find out conserved domains.
Phylogenetic analysis of NOTCH gene family
Multiple sequence was done by using bioinformatics tool MEGA 7 to obtain pairwise and multiple alignment of our sequence. MEGA 7 is used for manual sequence alignment and construct evolutionary tree. Maximum likelihood were used for tree construction. A separate instrument was used to measure molecular weight and isoelectric point. Different subfamilies were named according to their corresponding NOTCH homolog by phylogenetic analysis.
Gene structure analysis and detection of conserved motifs
Gene structure and display server are used to find the composition and positions of introns and exons and conserved elements of NOTCH gene.
The sequence of NOTCH protein was given as a query sequence and downloaded all target sequence in order to analyze the conserved motifs and domains of the protein sequence among (Pan Troglodytes), (Macaca mulatta), (Macaca nemestrina), (Pongo abelii) by using online database Pfam . This process is used to remove sequence that do not have conserved domain and motif. Position of chromosome, length of protein and genomic sequence was observed by using NCBI database. (http://pfam.janelia.org/)
Results and Discussion
NOTCH family has four members NOTCH1, 2,3and 4. NOTCH are involved in hair growth. NOTCH1 to 4 share high degree of structure similarity. NOTCH protein extracellular domain consist of variable number of epidermal growth factor that mediate interactions with ligands. It was originally identified in human leukemogenesis(Pancewicz and Nicot 2011). NOTCH can act as tumor suppressor or oncogenes are not clearly identified. It is also act as growth inhibitor in keratinocytes and small lung cancer. NOTCH1 gene can be observed by different tools to check their functions their conserved regions, introns exons, composition of elements and their evolutionary relation with their ancestors by using different tools such as MEME, MEGA 7, Gene structure and display server.
MEME
It is used to discover motifs of DNA, RNA and protein sequences. NOTCH1 gene protein sequence are put in MEME search box it can find their motifs the width of all the protein sequence are 50. The E value varies of each sequence.
Genome structure Display server: It was performed for the visualization of NOTCH1 gene composition, introns and exons and their conserved regions (Figure 2).
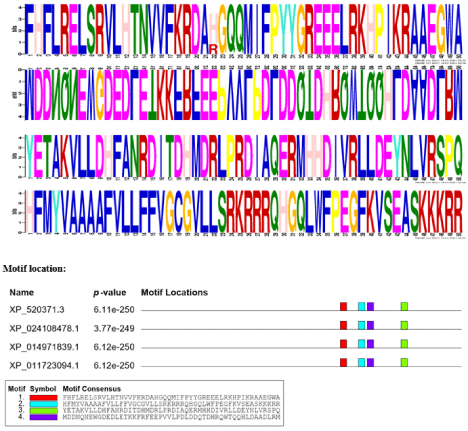
Figure 1:
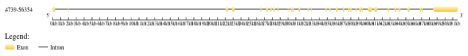
Figure 2:
MEGA7
Mega 7 tool is used to find the evolutionary relationship between different species. Diagram that describe the relation of organism, species from their common ancestors. Figure depict the molecular phylogenetic analysis by Maximum likelihood method. The analysis involved 11 amino acid sequence. All positions containing gaps and missing data were eliminated. Evolutionary analysis were conducted in Mega 7 (Figure 3).

Figure 3:
Serial cloner: It is used for the visualization and sequence analysis. It is also used in DNA cloning. It can used genomic sequence to find out the restriction site in genes. NOTCH1 gene mRNA and genomic sequence of different organisms are put in serial cloner to find the number of restriction sites in genes (Figure 4).
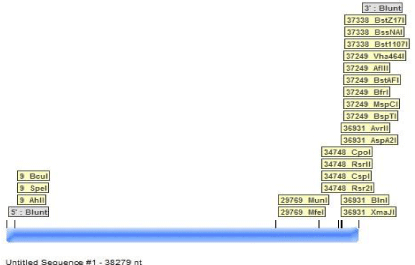
Figure 4:
Conclusion
Notch gene was first discovered in Drosophila (Egan, St-Pierre et al. 1998). NOTCH signaling pathway is important mediator of cell fate selection which are involved in epidermal appendage formation. Its four receptor play important role in cell differentiation in early development process. NOTCH protein share a high degree of domain topology. NOTCH signaling is mediated by intracellular domains which functions as transcription factor. Intracellular domains of NOTCH comprised different identified domains that have different functions such as regulating NOTCH activity (Gridley 2003). All members of NOTCH are involved in cancer including breast, melanoma, and leukemia, (Demarest, Ratti et al. 2008) pancreatic. The mutant form of NOTCH have been identified to increase NOTCH transcriptional activity. Their phylogenetic analysis depict the relation with their ancestors.
References
- Borggrefe T, Oswald A. "The Notch signaling pathway: transcriptional regulation at Notch target genes." Cellular and Molecular Life Sciences. 2009; 66: 1631-1646.
- Demarest R, et al. "It's T-ALL about Notch." Oncogene. 2008; 27: 5082-5091.
- Egan S, et al. “Notch receptors, partners and regulators: from conserved domains to powerful functions." Protein Modules in Signal Transduction. 1998; 273-324.
- Fleming RJ. Structural conservation of Notch receptors and ligands. Seminars in cell & developmental biology, Elsevier. 1998.
- Gridley T. "Notch signaling and inherited disease syndromes." Human molecular genetics. 2003; 12: R9-R13.
- Pancewicz J, Nicot C. "Current views on the role of Notch signaling and the pathogenesis of human leukemia." BMC cancer. 2011; 11: 1-7.
- Powell B, et al. "The Notch signalling pathway in hair growth." Mechanisms of development. 1998; 78: 189-192.