
Research Article
Austin J Plant Biol. 2015; 1(2): 1008.
Molecular Characterization of Bunt Resistance in Romanian Wheat Line F00628G34-M, Selected From a Triticale (Triticosecale) X Winter Bread Wheat (Triticumaestivum) Cross
Ciuca M*, Cristina D and Turcu AG
Department of National Agricultural Research, University of Development Fundulea, Europe
*Corresponding author: Matilda Ciuca, Department of National Agricultural Research, University of Development Fundulea, 915200, Calarasi, ROMANIA, Europe
Received: August 14, 2015; Accepted: November 23, 2015; Published: November 27, 2015
Abstract
Bunts are diseases produced by Tilletiacaries, T. laevis, T. controversa and T. secalis with severe effects on wheat yield and quality. Their importance has increased, especially in organic agriculture, where chemical treatments are forbidden. Rye is highly resistant to Tilletia sp. Breeders at NARDI Fundulea obtained a bunt resistant line F000628G34-M derived from a Triticale/2* wheat cross. Sixty eight randomly extracted F4 lines from a cross between F000628G34-M and susceptible wheat cultivarLiterawere phenotyped (using local populations of Tilletia sp.) and genotyped with 20 specific markers for 1 RS and 15 SSR markers located on 1AS. Chi square test showed significant deviation (P < 5%) from the expected Mendelian monogenic segregation, suggesting that the resistance gene is recessive or partially dominant and/or the resistance is affected by suppressing factors from wheat genome. Molecular markers assay proved a significant association of the bunt resistance inherited from the F00628G34-M line, with the 1A/1R translocation, suggesting that the bunt resistance gene originated from rye could be locatedin the region homeologous with the Glu-A3 locus and close to Xgwm1223 microsatellite locus. To our knowledge, this is the first time when it is proven that a bunt resistance gene is associated with the rye chromatin transfer to wheat.
Keywords: Wheat; Rye; Triticale; Bunt; Molecular markers; Tilletia sp
Introduction
Agriculture in Europe has been moving toward organic farming and with “low-inputs”, including reduced chemicals use. Reduction or lack of chemical seed treatments have lead to resurgence of many seed-borne diseases including bunts, which were previously controlled with chemicals [1]. In Romania, if untreated seeds are used, the incidence of bunt can reach up to 70-80% and the yield losses can reach up to 40% [2].
Wheat bunts are produced by Tilletia caries (DC) Tul, T. laevis (Wallr) Liro. (Common bunt) and T. controversa (dwarf bunt), but T. secalis (on rye) can also affect wheat. Although chemical treatments have been successfully used to control bunt, genetic resistance is the most economical and ecological way of control, and is the only acceptable approach in organic agriculture. This is why bunt resistance has become an important objective in wheat breeding. At present, have been identified more than 15 resistant genes to common bunt (Bt1 to Bt15) and in addition, the gene Bt-Z in Agropyroninter medium, the gene Bt-P in wheat variety PI 173438, a new gene in Blizzard and also, a QTL on 7B chromosome in McKenzie cv designated QCbt.spa-7B.1 [3-6]. The efficiency of resistance genes is different, but not very different from one area to another. In Romania the following genes have been identified with a good efficiency: Bt5, Bt8, Bt10, Bt11, Bt12, Bt13 [7].
Rye (Secalecereale L.) has already provided genes that proved useful in wheat breeding [8,9] and many cultivars that enjoyed a great success over time are carriers of translocations from rye, especially of some translocations in which the short arm of 1R chromosome is involved There are still many genes of interest for wheat improvement, not yet transferred, that are available in the rye genome, and that includes bunt resistance genes.
Phenotype observations conducted in the Czech Republic on 17 triticale cultivars showed presence of T. caries pathogen in a very low percent (2.4%) only on one triticale cultivar (Triamant) [10]. This result suggested that, due to the presence of rye chromatin, triticales are very resistant to common bunt attack. However, both rye and triticale were attacked by T. controversa (dwarf bunt). Similar results were reported by other researchers [4].
Triticale can be successfully used as a bridge for transferring useful rye genes to wheat [9]. The line F00628G34-M, created at NARDI Fundulea by crosses between triticale (Triticosecale) and wheat (Triticumaestivum), showed good resistance to bunt in artificial infections, both in tests from Romania, done at Fundulea [7] and Simnic [11] and in most locations of international tests from the European project “Tilletia Ring Test”. In the frame of this project, the F00628G34-M line was affected by Tilletia spp. at different levels, but considerably less than the susceptible entries (Austria -11.5%, Germany -15.5%, Croatia -race T7- 7.4%; race T9- 4.2%; race T17- 9.3%; race T02 – 4.7%) [12]. This line was previously characterized as carrying rye chromatin as a 1RS:1AL translocation, using hybridization techniques (GISH and FISH) at Research Institute of the Hungarian Academy of Sciences, Martonvásár, Hungary (Marta Molnar-Lang’s Department) by Constantina Banica (personal communication).
Marker Assisted Selection (MAS) has proved to be a very efficient method for increasing genetic progress and efficiency of breeding programs [13]. Starting from the necessity to obtain bunt resistant cultivars, taking into account the complexity of wheat genome, the difficulties of testing and selection of bunt resistant genotypes and the utility of molecular markers, this work proposes to look at the wheat genome with the help of molecular markers and to determine if the resistance to bunt in F000628G34-M line is associated with rye chromatin.
Biological material
The study was done on 68 F4linesrandomly extracted from the cross of the cultivarLitera, as susceptible parent, with the line F00628G34-M (Triticale/2* Wheat), as resistant parent.
Methods
Inoculation-teliospors
The teliospores were mixed with seed in a paper envelope, next was shaken by hand until the seed was evenly and visibly darkened with teliospores (performed under the guidance of Dr. Mariana Ittu).
Field tests
Inoculated seeds were planted on one meter long rows, using as susceptible cultivars Dropia and Literal. At maturity, the lines were classified in bunted and non-bunted (0 % common bunt).Infected spikes (where at least one grain was replaced by bunt balls) were counted and expressed as percentage from total number of spikes.
DNA isolation was made from leaves, using CTAB method [14]. PCR- All amplification reactions were carried out in a 25μl volume. We used 20 markers specific for rye and for the 1R short arm chromosome [15-21] and 15 SSR markers localized on the 1AS chromosome [22]. The sequences of REMS markers were kindly offered by Dr. Victor Korzun -KWS LOCHOW, Germany, and for marker Xgwm752 the sequence was obtained by MTA (Material Transfer Agreement) from Dr. Marion Roder. For the marker Xgwm1223 the sequence was obtained by MTA-from “TraitGenetics” company, Germany -Dr. Martin Ganal (Table 1). The PCR products were separated on 1.2-1.5-2-2.5% agarose for routine use, in 0.5X TBE buffer, stained with ethidium bromide and photographed under ultraviolet light with VilberLourmat system.
No.crt.
Name
Primerssequences
Location on chromosome
Markers for rye – specific localization on 1RS
1
F3 R3
F: GATCGCCTCTTTTGCCAAGA
R: TCACTGATCACAAGAGCTTG
Universal marker for rye
2
SCM9
F: TGACAACCCCCTTTCCCTCGT
R: TCATCGACGCTAAGGAGGACCC
1R, 1B
3
Bmac213
F: ATGGATGCAAGACCAAAC
R: CTATGAGAGGTAGAGCAGCC
1RS, 1H
4
Sec-1
F: CTATTAGTTCGAAAAGCTTATGA
R: GCATATGACTCAAATTATTTTTT
1R
5
IAG95
F: CTCTGTGGATAGTTACTTGATCGA
R: CCTAGAACATGCATGGCTGTTACA
1R
6
REMS1280
F: CAACGGCATGGAGTACCCT
R:GAAGTTAACCTGCGGGAACA
1R
7
REMS1135
F: GGCGTCCGTGTAGAGAGAGA
R: ACCTGATGCACCTCCAAAAG
1R, 3R, 7R
8
NOR
F: GCA TGT AGC GAC TAA CTC ATC
R: CCC AGT TTT CCA TGT CGC
1R
9
TSM12
F: CTGCGCACACATGAGTCAAT
R: ATGGAAGGCGAGAGTCCTTT
1R
10
TSM94
F: GGAGGCACCGAGATATTGAA
R: ACTTCCTCTAGGGCCGAAAC
1R
11
TSM106
F: AACGAACGGCAAGAACCTAA
R: GTCGGCTGCATCATCTCC
1R
12
TSM123
F: CCATCTCCCTCCTCCTGCTA
R: TGAACGACATGCTGTTAATATG
1R
13
TSM303
F: CAACACCGATAGGGTAGAGAGG
R: GATCATCGCCATCGTCATC
1R
14
TSM322
F:TGCCACACACAAACTTGACA
R:GCGAGATCGATGAAGAGAGC
1R
15
TSM325
F: CTGCACATGTCGCACCTC
R: AGGAGCCAAAGAAGCATCAA
1R
16
TSM422
F: GTCCTGCTGCTACTGTGCTG
R: CACACTCGCATCCTTTGCTA
1R
17
TSM556
F: GGGTAGGCAGAGGCCTAACTA
R:TACCCCTCTCCCTCCCTCT
1R
18
TSM608
F:AGGACGGGAAATAGGATGG
R: AACACATCCCCACTCTTGTTG
1R
19
TSM625
F: GTGTGAGAGAAAGCGAGAGAGAG
R: ATTTGTGATGCCGCTTATCC
1R
20
TSM690
F: CTGAATTGCTTTCGCGTTTT
R: GTAGTTTGGCCAGGCTGAAG
1R
Markers for 1AS chromosome
21
Xbarc1
F: GCGATGCTTTTGCCTTGTTTCAG
R: GCGGCCCCTTTGACTCTTCATAG
1A, 5A
22
Xbarc25
F: GCGGTGCATCAAGGACGACAT
R: GCGTAGTTCATCCATCCGTAAT1A, 3A, 4B
23
Xbarc148
F: GCGCAACCACAATGTATGCT
R: GGGGTGTTTTCCTATTTCTT
1A, 3A, 1D, 5B
24
Xbarc119
F: CACCCGATGATGAAAAT
R: GATGGCACAAGAAATGAT
1A, 1B, 1D
25
Xbarc263
F: GGAAGCGCGTCAGCACTAGGCAAC
R: GGCTTCTAGGTGCTGCGGCTTTTGTC
1A
26
Xbarc1048
F: ACG TGG TAA TTA GTT GGG AGT CTG TA
R: GCG AAG TCA AGA AGT GGG CTT TTC AAG AG
1A
27
Xgwm136
F:GACAGCACCTTGCCCTTTG
R:CATCGGCAACATGCTCATC
1A
28
Xgwm33
F:GGAGTCACACTTGTTTGTGCA
R:CACTGCACACCTAACTACCTGC
1A, 1B, 1D
29
Xgdm33
F:GGCTCAATTCAACCGTTCTT
R:TACGTTCTGGTGGCTGCTC
1A, 1B, 1D
30
Xwmc278
F: AAACGATAGTAAAATTACCTCGGAT
R: TCAAAAAATAGCAACTTGAAGACAT
1A
31
Xwmc336
F: GTCTTACCCCGCGATCTGC
R: GCGGCCTGAGCTTCTTGAG
1A, 1D
32
Xwmc818
F: TGAAGGGTGCGTGTGGTC
R: GCGTCGATTTTAATTTGATGATGG
1A, 1B
33
Xgwm1223
Material Transfer Agreement (Dr. Martin Ganal)
1R,1A, 1D
34
Xgwm750
Material Transfer Agreement (Dr. Marion Roder)
1R, 1A
35
PSP2999 (Glu-3)A
F: TCCCGCCATGAGTCAATC
R: TTGGGAGACACATTGGCC1A
Table 1: Molecular markers used in this study and their specificity.
As positive control for rye chromatin, we used DNA from Secalecereale “Harkowskaya”, from wheat cultivar Liman (which carries rye chromatin) and from another wheat genotype which carries the 1RS:1BL translocation Ludogoria (seeds were kindly provided by Dr. Elena Todorovska, ABI, Sofia, Bulgaria).
For nested PCR (F3/R3 - Xgwm1223), the final reaction was done in a 25 μl final volume using 3 μl of PCR product obtained with “universal marker for rye” F3/R3 [14]. We used 1U Taq DNA polymerase (Promega) and 0,2 &muM primers. Association between markers and level of bunt attack was estimated using Chi square test, interactive software [23].
Results
Phenotype observations
Phenotype observations made in 2010 for the F4 generation on 68 progenies lines, showed 18 progenies lines non-bunted and 50 progenies lines bunted, the percentage of bunted ears varying between 9 and 80 percent, whilst the susceptible parent, Literal showed 65 percent of bunted ears. We also observed partially bunted spikes and seeds. The phenotype segregation suggests that the bunt resistance gene transmitted from F00628G34-M manifested as recessive or partially dominant gene. However, Chi square test suggests significant deviation (P < 5%) from the expected Mendelian monogenic segregation (Table 2). This might be explained by repression of resistance by certain factors from wheat genome.
Non-bunted lines
Bunted lines
Total
Observed
18
50
68
Expected (segregation ratio 1,75:0,5:1,75)
29,75
8,5+29,75
68
O-A
-11,75
+11,75
χ2 =8,25
P= 0.00407468
Degrees of freedom = 1
Table 2: Chi-squared test for bunt attack in 68 progenies of the cross F00628G34-M/Literal.
The characterization of rye translocation present in F00628G34-M line
Molecular analysis with specific markers for rye chromatin offered some information about 1RS:1AL translocation present in F00628G34-M line. The marker SCM9 [19] that make difference between 1RS:1AS translocation (220bp PCR product) and 1RS:1BS translocation (200bp PCR product), confirmed the presence of 1RS:1AS translocation in F00628G34-M (Figure 1).
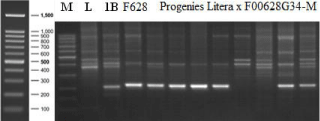
Figure 1: Electrophoresis pattern of SCM9 marker. M-DNA Ladder (100bp);
L-cv Literal; 1B-Ludogoria (1R/1B translocation); F628 - F00628G34-M line.
The molecular marker STS-IAG95 [15] showed a PCR product of 1150bp (Figure 2) in the line F000628G34-M. Based on previously reported results [15], with this marker one can obtain either 1050bp or 1150bp PCR product, the first being associated with “Petkus” rye type and the second PCR product (1150bp) with “Insave” rye type.
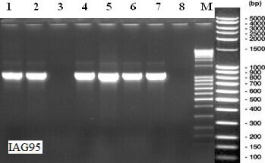
Figure 2: Electrophoresis pattern of IAG95 marker: 1-cv Liman (1R
translocation); 2-F00628G34-M; 3-cv Literal; 4-8 – progenies (Literal x
F00628G34-M); M- DNA ladder 100bp extended (Roth).
This locus is associated with a gene for resistance to Blumeriagraminis (Pm8 for “Petkus” type and Pm17 for “Insave” type). Because we obtained an 1150bp product, the rye chromatin from our line is INSAVE type, and probably contains the Pm17 powdery mildew resistance gene.
The 1RS arm chromosome carries a locus for secaline (ω secaline and γ secaline). Using the PCR marker for Sec-1 locus (ω secaline) [20] results in 2 PCR products (1.5kb and 0.7kb), but in our case we obtained the 1.5kb product and another 1.2kb product (Figure 3). Apparently, the Sec-1 locus showed polymorphism, presented by a PCR product of about 1200bp in our line. Maybe, this polymorphism is determined by the specificity of Insave rye chromatin present in the line F000628G34-M.
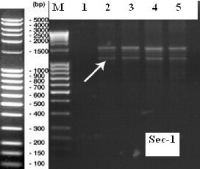
Figure 3: Electrophoresis pattern for Sec-1 locus. M-DNA ladder 100bp
extended (Roth); 1-cv Litera; 2-F00628G34-M; 3,4,5-progenies. The arrow
mark the 1,2 kb PCR product.
The association of bunt resistance with rye chromatin inline F000628G34-M based on molecular markers assays. Of all 35 markers tested in this study, only 12 markers showed polymorphism between parents. The results of chi-square test for these markers are presented in (Table 3). The results for the IAG95 were not included because its sequence has a transposable nature [24].
Marker Name
Location on
chromosome
Chi-squared test- Probability (for 68 F4 lines)
Chi-squared test- Probability (for only the homozygous lines)
F3 /R3
Rye-marker
0.00094212
0.00004733
SCM9
1R, 1B
0.00958200
0.00028462
Sec-1
1R
0.00222864
0.00002873
TSM106
1R
0.00958200
0.00028462
TSM123
1R
0.00958200
0.00028462
Xbarc263
1A
0.00012177
0.00010338
Xbarc1048
1A
0.00001093
0.00000832
Xgwm136
1A
0.00002097
0.00004733
Xgwm1223
1R,1A,1D
0.00036605
0.00004733
PSP2999 (Glu A3)
1A
0.00006456
0.00004733
Xwmc818
1A, 1B
0.00012177
0.00010338
Table 3: Association of molecular markers to bunt resistance.
Combined analysis of results obtained with molecular markers for Sec-1 locus and for Glu-A3 locus (PSP2999) permitted to distinguish heterozygous genotypes [25].
Both the presence of the universal marker for the rye chromatin and of four other specific markers for the 1R chromosome, and the absence of 6 specific markers for the 1A chromosome proved a significant association of the bunt resistance inherited from the F00628G34-M line, with the 1A/1R translocation. This proves that bunt resistance identified in the F00628G34-M line is associated with the presence of rye chromatin.
To our knowledge, this is the first time when it is proven that a bunt resistance gene is associated with the rye chromatin transfer to wheat. Molecular assays suggest a possible location of the bunt resistance gene in F00628G34-M line, on the 1RS chromosome, in the homeologous region with the Glu-A3 locus and close to Xgwm1223 microsatellite locus. Therefore the bunt resistance gene present in F00628G34-M is different from other known resistance genes. The markers association with the bunt resistance gene was more significant on homozygous genotypes.
For the transfer of this gene in other genotypes, we proved that the marker assisted selection can be used, the most indicated markers being Xbarc1048, Xgwm136 or PSP2999, whose absence notifies the presence of rye chromatin where the gene is localized.
Also, with the help of molecular markers (by nested - PCR with markers (F3/R3 / Xgwm1223), we showed that the 1RS:1AL translocation present in the line F000628G34-M has some chromosome rearrangements, by wheat chromatin insertion. This result was suggested by the polymorphism obtained using the PCR product (F3/R3) as DNA template for Xgwm1223 marker amplification, which was similar with PCR products by Xgwm1223 when DNA template came from genotypes without rye chromatin (Figure 4). This region could be heterogenic and can influence the molecular marker assay for association with resistance gene to bunt.
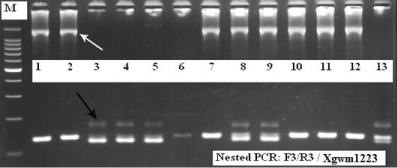
Figure 4: Electrophoresis pattern of nested PCR (F3R3/wms1223). M- DNA
ladder 100bp; 1. F00628G34-M; 2-12 progenies; cv Literal. The white arrow
mark the rye chromatin (F3R3 PCR product). The black arrow mark the
Xgwm1223 PCR product.
Discussion
The lower than expected number of non-bunted genotypes could be explained by incomplete gene expression or by suppression of the resistance gene from rye by modified/inhibitor factors from wheat genome. Similar results for disease resistance genes transferred from rye genome have already been reported [26].
The slow progress in learning about the genetic resistance to bunt is explained by a series of diseases characteristics [4]. In most of the cases, not all the plants with the susceptible genotypes are attacked, even if a good artificial infection is obtained. The resistant genotypes are rarely completely immune. Different authors define as resistant the genotypes that are attacked at a level less than 5 to 10%.
The existence of modifying genes can influence the infection level. Many of the resistance genes present a partial dominance. Additionally, the level of bunt infection is strongly influenced by the autumn environmental conditions, which makes the heritability relatively low.
All these aspects, but especially the incomplete expression of susceptibility or resistance alleles, distort the phenotypic data of resistance to bunt in mapping populations, and this is an obstacle in the determination of genetic distance between resistance gene and molecular markers. This problem could be partially solved if the study is realized on homozygous recombinant lines, preferably doubled haploid lines, which permit the phenotyping of the same genotype for more years to establish more clearly the resistance classes. When these kinds of populations are not available, the solution is a qualitative determination of the associations between resistance genes and markers using Chi-square test, without estimating the genetic distance.
The difficulties which remain for the study of genetic resistance to bunt can also be found out in breeding bunt resistant cultivars. Moreover, the low agronomic performances of most of the known resistance sources make the association of the resistance to bunt with some unfavorable agronomical traits difficult to deal with, even after several crossing cycles. Compared to other wheat diseases, all these problems make the wheat selection for bunt resistance to be considered one of the most difficult. The determination, even only at the qualitative level, of some molecular markers association with bunt resistance genes could accelerate the introduction of these genes in wheat genotypes with good agronomic traits. That is why, for obtaining wheat cultivars with high-performance and resistance to bunt, the use of molecular markers associated to bunt resistance genes is of great importance.
Conclusion
The bunt resistance identified in the F00628G34-M line is associated with the presence of rye chromatin.
For the transfer of this gene in other genotypes we proved that the marker assisted selection can be used, the most indicated markers loci being Xbarc1048, Xgwm136 or PSP2999 (GluA3), whose absence notifies the presence of rye chromatin in the region where the gene is located. The Xgwm1223 marker locus, which produces specific products for the rye translocation, can also be used.
The bunt resistance gene present in F00628G34-M is different from other known resistance genes.
References
- Lammerts van Bueren ET, Østergård H, Goldringer I, Scholten O. Plant breeding for organic and sustainable, low-input agriculture: Dealing with genotype-environment interactions. Euphytica. 2008; 163: 321-322.
- Cota LC, Botez C, Grigoras M, Curticiu D. Screening for resistance to artificial infection by common bunt (Tilletia caries and Tilletiafoetida) in F2 populations of wheat (TriticumaestivumL.). Bull Univ Agric Sci Vet Med (USAMV). 2009; 66: 24-31.
- Hoffmann JA, Metzger RJ. Current status of virulence genes and pathogenic races of the wheat bunt fungi in the northwestern USA. Phytopathology. 1976; 66: 657-660.
- Goates BJ, Wilcoxson RD, Saari EE. Common bunt and dwarf bunt. In: Bunt and Smut Diseases of Wheat: Concepts and Methods of Disease Management. CIMMYT Mexico City. 2014; 3: 201-207.
- Wang S, Knox RE, DePauw RM, Clarke FR, Clarke JM, Thomas JB. Markers to a common bunt resistance gene derived from 'Blizzard' wheat (Triticumaestivum L.) and mapped to chromosome arm 1BS. Theor Appl Genet. 2009; 119: 541-553.
- Knox RE, Campbell HL, Depauw RM, Gaudet D, Puchalski B, Clarke FC. DNA markers for resistance to common bunt in ‘McKenzie’ wheat. Canadian Journal of Plant Pathology. 2013; 35: 328-337.
- Ittu M, Saulescu NN, Ittu G. 2006 - Latest in breeding for resistance to common bunt in Romania. Czech Journal of Plant Breeding. 2006; 42: 15.
- Kumar S, Mittal1 RK, Dhiman R, Gupta D. Assessment of Triticale (Triticosecale) X Bread Wheat (TriticumAestivum) Genotypes for Drought Tolerance Based on Morpho-Physiological, Grain Yield and Drought Tolerance Indices Under Non-Irrigated and Irrigated Environments. International Journal of Food Science, Nutrition and Dietetics. 2014; 1-7.
- Saulescu NN, Ittu G, Ciuca M, Ittu M, Serban G, Mustatea P. Transferring useful rye genes to wheat, using triticale as a bridge. Czech J Genet Plant Breed. 2011; 47: 56-62.
- Dumalasová V, Bartoš P. Reaction of wheat, alternative wheat and triticale cultivars to common bunt. Czech J Genet Plant Breed. 2010; 46: 14-20.
- Oncică, F, Saulescu NN. Potentially new sources of genes for resistance to common bunt (Tlletia spp.) in winter wheat (Triticumaestivum L. Proceedings of the Romanian Academy, Series B- Chemistry, Life Sciences Geosciences. 2008: 97-100.
- Săulescu N, Ittu G, Giura A, Ciucă M, Mustătea P, Ittu M. Diversific area bazeigenetice ca fundament al progresuluiî namelio rare agrâului. Analele Institutului INCDA Fundulea. 2010; 8: 1-14.
- Miedaner T, Korzun V. Marker-assisted selection for disease resistance in wheat and barley breeding. Phytopathology. 2012; 102: 560-566.
- Sagai-Maroof MA, Soliman K, Jorgensen RA, Allard RWM. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location and population dynamics. PNAS. 1984; 81: 8014-8018.
- Roder MM, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, et al. A microsatellite map of wheat. Genetics. 1998; 149: 2007-2023.
- Kofler R, Bartos J, Gong L, Stift G, Suchankova P, Simkova H, et al. Development of microsatellite markers specific for the short arm of rye (Secalecereale L.) chromosome 1. Theor Appl Genet. 2008; 117: 915-926.
- Katto MC, Endo T R, Nasuda S. A PCR- based marker for targeting small rye segments in wheat background, Genes & Genetic Systems. 2004; 4: 245- 250.
- Mohler V, Hsam SLK, Zeller FJ, Wenzel G. An STS marker distinguishing the rye-derived powdery mildew resistance alleles. Plant Breed. 2001; 120: 448-450.
- Saal B, Wricke G. Development of simple sequence repeat markers in rye (SecalecerealeL.). Genome. 1999; 42: 964-972.
- Shimizu Yu, Nasuda S, Endo TR. Detection of the Sec-1 locus of rye by a PCR-based method. Genes & Genetic Systems. 1997; 72: 197-203.
- Wang L, Genying L, Pea JR, Xia X, Zhonghu H. Development of STS markers and establishment of multiplex PCR for Glu-A3 alleles in common wheat (Triticumaestivum L.). Journal of Cereal Science. 2010; 51: 305-312.
- Graine Genes. A database for Triticeae and Avena.
- Preacher KJ. Calculation for the chi-square test: An interactive calculation tool for chi-square tests of goodness of fit and independence [Computer software]. 2001.
- Gyawali YP, Nasuda S, Endo TR. A cytological map of the short arm of rye chromosome 1r constructed with 1R dissection stocks of common wheat and PCR-based markers. Cytogenet Genome Res. 2010; 129: 224–233.
- Devos KM, Bryan GJ, Collins AJ, Stephenson P, Gale MD. Application of two microsatellite sequences in wheat storage proteins as molecular markers. Theoretical and Applied Genetics. 1995; 90: 247-252.
- McIntosh RA, Zhang P, Cowger C, Parks R, Lagudah ES, Hoxha S. Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. TheorAppl Genet. 2011; 123: 359-367.