
Research Article
Austin J Plant Biol. 2016; 2(1): 1014.
Chemotaxonomic Significance of Flavonoids in Some Species of Galium (Rubiaceae) from Libya
Moubasher H¹*, Abd E¹-Ghani M¹, Al-Wakeel S¹ and Bahoor A¹
¹Department of Botany and Microbiology, Cairo University, Egypt
²Faculty of Science, Al-Marqeb University, Libya
*Corresponding author: Moubasher H, Department of Botany and Microbiology, Faculty of Science, Cairo University, Giza 12613, Egypt
Received: July 22, 2016; Accepted: September 06, 2016; Published: September 12, 2016
Abstract
The flavonoid profiles of five Libyan Galium L. (Rubiaceae) species collected from different localities and habitats were investigated. Fourteen flavonoid compounds were isolated and identified using the direct comparison of chromatographic and UV spectral analyses with standard samples. The flavone compounds were identified as apigenin (1) and its 7-glycoside (2); luteolin 7-diglycoside (3), diosmetin (4) and its 7-monoglycoside (5), as well as its 7-diglycoside (6). In addition, the detected flavonol compounds were kaempferol 3-glycoside (7), 3-diglycoside (8) and 3,7-diglycoside (9); quercetin (10) and its 3-glycoside (11), 3-rutinoside (12), 3,7-diglycoside (13), and 3-diglycoside- 7-glycoside (14). The chemotaxonomic relationships of the studied species of Galium and their significance were also discussed.
Keywords: Chemosystematics; Galium; Rubiaceae; Flavonoid glycosides; biochemistry; Multivariate analysis
Introduction
Galium L. is one of the largest genera of Rubieae (Rubiaceae) with more than 400 species included into 16 sections containing annual and perennial herb that are distributed in temperate and tropical regions of the world [1,2]. Certain species of Galium are found even in the Arctic zone or high elevations on tropical mountain ranges. Galium itself is problematic taxonomically, because taxa from different sections exhibit similar habit, many species are widely distributed and polymorphic, and species groups often are poorly differentiated both morphologically and geographically [3].
The flavonoid chemistry of Galium have been studied, and reported to contain predominantly flavones and flavonol aglycones and their glycosides. Earlier study by [4] isolated rutin and a mixture of flavonoids and diosmetin from Galium palustre L. Also, the glycosides of quercetin, luteolin, apigenin and kaempferol were isolated from Galium aparine [5,6]. Reported that Galium has long been known to contain substantial amounts of anthraquinones, with the roots being especially rich sources of these secondary metabolites. Bedstraw species, including G. mollugo, contain mollugin [7], flavonoids [8], coumarins, phenolic acids, and iridoid glucosides [9]. Recently, two new flavonoids; diosmetin glycoside and biflavone, were isolated and identified in G. verum L., in addition to isorhamnetin and its glycosides, kaempferol, quercetin, diosmetin and its glycosides [10,11].
Bedstraw species, including G. mollugo, also contain mollugin [7], flavonoids [8], coumarins, phenolic acids, and iridoid glucosides [12,13]. Some of these compounds have allelopathic, fungistatic, or repellent effects, and may also be used to flavour food or wine [14]. Recently, extracts from this plant were evaluated for their anti-cancer and anti-malarial activities and for their ability to inhibit HIV–1 reverse transcriptase, but initial results showed no activity [15].
The flora of Libya is not rich in the number of species; however, the Al-Jabal Al-Akhdar Mountain landscape comprises the richest vegetation and the highest number of species known from Libya [16]. The geographical affinity of the flora is mainly East Mediterranean rather than neighboring regions of North Africa [17,18].
In Libya, Rubiaceae is one of the eight species-rich families which represented by 50 genera and 90 species [19]. The genus Galium is represented by 10 species; G. mollugo L., G. spurium L., G. aparine L., G. tricornutum Dandy, G. verrucosum Huds., G. parisiense L., G. setaceum Lam., G. recurvum Req. ex DC., G. cossonianum Jafri, and G. murale (L.) All.
As from the beginning of 1990s, the increasing interest of molecular approach in taxonomic studies, especially those based on nucleic acids sequences, a remarkable decrease in number and importance of investigations dealing with chemotaxonomic evidence. Therefore, some authors urged this trend, and recommended research work on non-molecular evidences [20].
There is often confusion between different samples of this genus in herbariums and so the aim of this study is to develop a method to find characters providing data of both taxonomical and pharmaceutical use. No phytochemical studies were reported so far from Galium species in North African countries. The present work aims to establish foliar flavonoid patterns of five Galium species from Libya, in an attempt to determine chemical affinities among species and compare the results obtained with affinities indicated by evidences from morphology.
Materials and Methods
Plant material
Table 1 shows locations, types of habitats, dates of collection, GPS coordinates of localities, elevation and number of populations that were examined for the selected 5 species of Galium. Aerial parts of all plants were collected from natural populations. For each species at least two populations from distant habitats were studied. Altogether, 80 specimens were collected from their natural habitats in different locations of Libya. Voucher specimens of the studied species were deposited at the herbarium of Cairo University (CAI).
Species
Localities
Habitats
Elevation (m ASL)
GPS coordinates
Galium aparine L.
Gasr Libya (S4)
Mountainous areas
280.7
32° 37’N, 21° 23’E
Shahat Ruins (S1)
Mountainous areas
560.2
32° 50’N, 21° 55’E
Wadi Qaam (S12)
Coastal wadis
20.7
32° 28’N, 14° 25’E
G. murale L.
Wadi Elkouf (S5)
Inland desert wadis
264.6
32° 41’N, 21° 33’E
Sharshara (S8)
Canal banks
327.4
32° 27’N, 13° 37’E
G. setaceum Lam.
Wadi Derna (S6)
Inland desert wadis
85.3
32° 42’N, 22° 36’ E
G.tricornutum Dandy
Lamluda (S2)
Farmlands
barley fields
668.1
32° 44’N, 22° 06’E
Almansoura (S3)
Farmlands
NA
32° 50’N, 21° 55’E
Stwa (S7)
Farmlands
566.9
32° 49’N, 22° 09’E
G. verrucosum Huds.
Wadi Qaam (S11)
Coastal wadis
20.7
32° 28’N, 14° 25’E
Almansoura (S9)
Farmlands
NA
32° 50’N, 21° 55’E
Wadi Derna (S10)
Inland desert wadis
85.3
32° 42’N, 22° 36’ E
Table 1: List of studied species with binomials, habitats, localities, collection dates and GPS coordinates of localities. Figures between parentheses refer to the population numbers. NA=Not Available.
Biochemical procedures
The whole plant of Galium was dried in the shade and ground. The powder was extracted with 70% MeOH three times at room temperature and evaporated under reduced pressure. The aqueous layer was stored in a refrigerator until needed to test the presence of flavonoids in different samples.
The separation of flavonoid mixture was detected by spotting all the flavonoid extracts using one-dimensional paper chromatography on Whatman No. 3mm along with the standard samples, water, 15% acetic acid (acetic acid: water; 15:85) and BAW (n-butanol: acetic acid: water; 4: 1: 5) upper phase as solvent systems. After drying the paper chromatograms, the developed spots were examined under UV light at a wave length of 365nm using an Ultraviolet Lamp (Model, UV GL-25) then in the presence of ammonia fumes.
The qualitative analysis of the crude plant extracts containing complex mixture of flavonoid compounds was carried out on Whatman No. 3mm chromatographic paper, using the solvent system BAW in the first run and 15% acetic acid in the second. The chromatograms should be dried in the hood between the first and the second chromatographic runs. All the flavonoid spots on the dried chromatograms were detected under UV light with and without the presence of ammonia.
The separation of a flavonoid mixture was achieved by elution techniques using one-dimensional paper chromatography on Whatman No. 3mm. The used solvent system in purification of flavonoid mixture was selected after preliminary one-dimensional runs in different systems to observe which has effectively separated the mixture. The used solvent systems were BAW (n-butanol: acetic acid: water; 4: 1: 5) upper phase; 15% acetic acid (acetic acid: water; 15:85) and water. The crude plant extract was applied as a band at 10cm from the top of the chromatograms and developed in the selected solvent. UV–detectable bands observed on dried chromatograms with and without exposure to ammonia fumes were cut out and eluted with 95% methanol. Each methanolic extract was concentrated by evaporation on rotary evaporator. Additional purification of the isolated flavonoids was carried out until a pure flavonoid compound was obtained.
Ultra Violet (UV) spectral analyses were performed according to standard procedures performed by Mabry et al. [21] and Mabry and Markham [21].
The diagnostic reagents used for the UV spectral measurements of the isolated flavonoid compounds were: Sodium Methoxide (NaOMe), Aluminium Chloride (AlCl3), Hydrochloric Acid (HCl), Sodium Acetate (NaOAc), and Boric Acid (H3BO3). The absorption spectra were measured in methanolic solution against methanolic blank using automatic recording Spectrophotometer (UV–3101 PC, UV-VIS-NIR) scanning Spectrophotometer using standard Quartz cuvettes of 1cm path length. A fresh stock solution of 0.1mg pure flavonoid in about 10ml spectral methanol was prepared and adjusted on the spectrophotometer so that the major absorption peaks between 240 and 460nm. Then, the following steps were carried out:
- The MeOH spectrum was recorded at normal scan speed, using 2-3ml of the fresh stock solution.
- The NaOMe spectrum was recorded immediately after addition of 3 drops of NaOMe stock reagent to the solution used for step (1). After 5 minutes, the spectrum was rerun to check for any decomposition in the compound, this solution was discarded,
- The AlCl3 spectrum was recorded immediately after the addition of 6 drops of AlCl3 to 2-3ml of the fresh stock methanol solution,
- The AlCl3/HCl spectrum was recorded immediately after addition of 3 drops of stock HCl reagent to the cuvette containing AlCl3 used for step (3), the solution was then discarded,
- The NaOAc spectrum of the flavonoid was determined by adding enough anhydrous NaOAc to the cuvette containing 2-3ml fresh stock solution of the flavonoid. After shaking, recording out within two minutes and then after 5-10 minutes to check for any decomposition of the compound,
- The NaOAc/H3BO3 spectrum was recorded after addition of anhydrous H3BO3 to the cuvette which contained NaOAc used for step (5).
Multivariate analysis, using the species analyzed as Operational Taxonomic Units (OUT) and the flavonoids as characters, was carried out with PAST ver. 2.11 [22]. The resulting UPGMA dendrogram is shown in (Figure 1). Principal Components Analysis (PCA) was performed sing MVSP ver. 3.1 [23] and the resulting biplot is shown in (Figure 2).
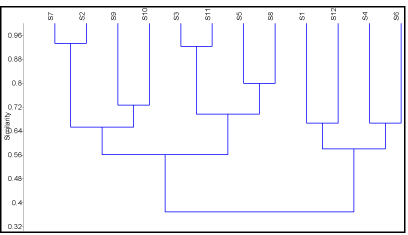
Figure 1: Dendrogram of Galium sp. based on flavonoid distributions and UPGMA clustering method. A, B and C are the three separated groups. For species
abbreviations, see (Table 1).
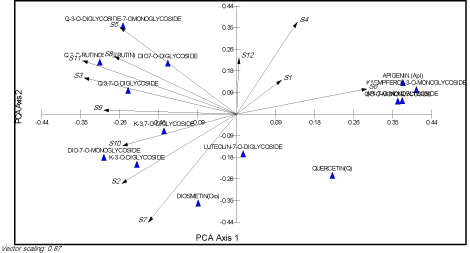
Figure 2: PCA scatter biplot showing the relations between the separated compounds and the studied species (S1-S10). A, B and C are the separated groups in
(Figure 1).
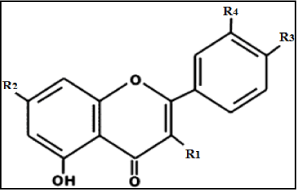
table 3: Chemical structure of isolated flavonoids from the five investigated
Galium species.
Results and Discussion
According to recent modifications to the classification of Galium [23], the 5 studied species are included in two sections: Sect. Kolgyda (Galium murale L., G. tricornutum Dandy, G. aparine L. and G. verrucosum Huds.) and Sect. Jubogalium (G. setaceum Lam.).
Fourteen flavonoid compounds were detected in the five Galium species collected from different locations of Libya. Identification of the isolated flavonoids was mainly based on the direct comparison of chromatographic and UV spectral studies with standard samples. The present data clearly established that the investigated species have a simple and highly substituted flavone and flavonol compounds including flavonoid, aglycones, glycosides and methylated compounds. The isolated flavonoids based on six flavones and eight flavonols. The flavone compounds identified were apigenin (1) and its 7-glycoside (2), luteolin 7-diglycoside (3), diosmetin (4) and its 7-monoglycoside (5), as well as its 7-diglycoside (6). In addition, the detected flavonol compounds were kaempferol 3-glycoside (7), 3-diglycoside (8) and 3,7-diglycoside, (9) quercetin, (10) and its 3-glycoside (11), 3-rutinoside (12), 3,7-diglycoside (13), and 3-diglycoside-7-glycoside (14).
Identification of the isolated flavonoids
Paper chromatography: The colour reaction of the aglycone (apigenin) appeared as purple spot on 1D and 2D paper chromatogram under UV light and with ammonia fumes, whereas the aglycone: quercetin showed yellow spot with UV light, that changed to bright yellow when fumed with ammonia (Table 2). The methylation of C-4` position with and without glycosidation at C-7 appeared as purple spots on paper chromatogram upon exposure to UV light and after fumed with ammonia. Additionally, most of the other glycosides appeared as purple spots with UV light and changed to yellow with ammonia fumes.
No.
Compounds
Rf value ×100
Colour reaction
BAW
15% acetic acid
UV
UV/NH3
1
Apigenin
92
2
P
P
2
Apigenin-7-O-glycoside
47
27
P
YG
3
Luteolin-7-O-diglycoside
33
29
P
Y
4
Luteolin-4`-O-Me (Diosmetin)
76
4
P
dP
5
Diosmetin-7-O-glycoside
34
40
P
dP
6
Diosmetin-7-O-diglycoside
20
63
P
dP
7
Kaempferol-3-O-glycoside
57
35
P
Y
8
Kaempferol-3-O-diglycoside
35
60
P
Y
9
Kaempferol-3,7-O-diglycoside
25
70
P
Y
10
Quercetin
69
3
Y
brit Y
11
Quercetin-3-O-glycoside
56
35
P
Y
12
Quercetin-3-O-rutinoside (Rutin)
33
46
P
Y
13
Quercetin-3,7-O-diglycoside
24
72
P
Y
14
Quercetin-3-O-diglycoside-7-O-glycoside
16
81
P
YG
Table 2: Paper chromatographic data of the isolated flavonoid compounds. Y=yellow; P=purple; dp=dull purple; YG=yellow green; britY=bright yellow.
The characteristic Rf values of each compound on 1D paper chromatograms in different solvent systems (Table 2) indicated the position of glycosides attachment. The mobility of a glycoside is closely correlated with the number and position of sugar substitution. Increasing glycosylation at C-3 position decreased Rf value in BAW and increased the mobility in 15% acetic acid relative to its aglycone. The 3,7-diglycosidation behave like the 3-glycosidation, but moved very slower in BAW and faster in 15% acetic acid. In addition, increasing glycosidation at C-7 position with methylated C-4` of luteolin showed faster movement in 15% acetic acid and lower Rf value in BAW relative to its aglycone (diosmetin).
Ultra Violet spectra (UV): The UV spectral data of isolated flavonoid compounds are shown in (Table 3). In addition, the chemical structures of the isolated flavone and flavonol compounds are illustrated in (Table 4).
Compound
MeOH
NaOMe
AlCl3
AlCl3/HCl
NaOAc
NaOAc/H3BO3
(1) Apigenin
336
392
382
381
340
337
268
325
345
342
302 sh
269
. . .
275
301
300
269
. . .
. . .
. . .
277
278
. . .
. . .
(2) Apigenin-7-O-glycoside
331
380
381
380
385
337
288 sh
348 sh
347
340
352 sh
265
265
302
296
295
265
. . .
. . .
270
272
274
. . .
. . .
(3) Luteolin-7-O-diglycoside
347
392
428
386
407
369
265 sh
263
328
357
365 sh
260
250
. . .
296 sh
295 sh
265
253 sh
. . .
. . .
275
272
257 sh
. . .
(4) Luteolin-4`-O-methyl (Diosmetin)
344
382
384
355
343
342
289
270
360
295
271
270
270
. . .
295
276
. . .
. . .
. . .
. . .
273
. . .
. . .
. . .
(5) Diosmetin-7-O-glycoside
343
372
382
383
340
342
265
300 sh
360
355
264 sh
264 sh
250
266
292 sh
293 sh
255
254 sh
. . .
. . .
272
269
. . .
. . .
(6) Diosmetin-7-O-diglycoside
343
370
381
383
340
342
267
300 sh
360 sh
360 sh
282 sh
264 sh
251
266
293 sh
293 sh
264 sh
253
. . .
. . .
270
269
253
. . .
(7) Kaempferol-3-O-glycoside
352
403
397
395
372
352
295 sh
326
350
346
302
295
265
274
304
303
275
265
. . .
. . .
274
274
. . .
. . .
(8) Kaempferol-3-O-diglycoside
352
406
403
400
393
352
320 sh
325
358
346
312
320 sh
267
278
304
302
277
295 sh
. . .
. . .
276
275
. . .
266
(9) Kaempferol-3,7-O-diglycoside
352
392
394
394
400
352
315 sh
350 sh
353
348
358 sh
300 sh
263
300 sh
300 sh
300 sh
265
268
. . .
268
270
270
. . .
. . .
(10) Quercetin
372
419
452
430
373
386
306 sh
331
337
362
256
260
256
. . .
271
267
. . .
. . .
(11) Quercetin-3-O-glycoside
357
405
428
400
390
377
295
326
301 sh
358
271
288 sh
266 sh
267
273
29 sh
. . .
260
255
. . .
. . .
269
. . .
. . .
(12) Quercetin-3-O-rutinoside (Rutin)
358
417
429
400
363
380
306 sh
328
275
364
267
263
259
273
. . .
299
. . .
. . .
. . .
. . .
. . .
270
. . .
. . .
(13) Quercetin-3,7-O-diglycoside
354
430
433
400
437
380
266 sh
277
300 sh
356
385 sh
282 sh
255
. . .
273
298
262
265
. . .
. . .
. . .
268
. . .
. . .
(14) Quercetin-3-O-diglycoside-7-O-glycoside
356
410
438
400
418
375
295 sh
269
330 sh
364 sh
375 sh
262
268 sh
. . .
298 sh
298 sh
297 sh
. . .
258
. . .
273
268
263
. . .
Table 3: UV spectral data of isolated flavonoid compounds (λmax, nm); sh=shoulder.
No.
Compound
R1
R2
R3
R4
1
Apigenin
H
OH
OH
H
2
Apigenin-7-O-glycoside
H
O-monoglycosyl
OH
H
3
Luteolin-7-O-diglycoside
H
O-diglycosyl
OH
OH
4
Luteolin-4`-O-Me (Diosmetin)
H
OH
O-CH3
OH
5
Diosmetin-7-O-glycoside
H
O-monoglycosyl
O-CH3
OH
6
Diosmetin-7-O-diglycoside
H
O-diglycosyl
O-CH3
OH
7
Kaempferol-3-O-glycoside
O-monoglycosyl
OH
OH
H
8
Kaempferol-3-O-diglycoside
O-diglycosyl
OH
OH
H
9
Kaempferol-3,7-O-diglycoside
O-monoglycosyl
O-monoglycosyl
OH
H
10
Quercetin
OH
OH
OH
OH
11
Quercetin-3-O-glycoside
O-monoglycosyl
OH
OH
OH
12
Quercetin-3-O-rutinoside (Rutin)
O-rhamnoglucosyl
OH
OH
OH
13
Quercetin-3,7-O-diglycoside
O-monoglycosyl
O-monoglycosyl
OH
OH
14
Quercetin-3-O-diglycoside-7-O-glycoside
O-diglycosyl
O-monoglycosyl
OH
OH
Table 4: Chemical structure of isolated flavonoids from the five investigated Galium species.
Distribution patterns of the isolated flavonoids
The qualitative and quantitative analyses of the 14 flavonoids among the five Galium species that collected from different locations of Libya are represented in (Table 5). The flavonoid patterns between the Galium species showed a predominant flavonol glycoside being quercetin-3-rutinoside (12). However, the other compounds were variable in quality, quantity, and in their distribution among the investigated samples.
Compound
G. setaceum
G. aparine
G. tricornutum
G. verrucosum
G. murale
S6
S4
S1
S12
S7
S2
S3
S9
S10
S11
S5
S8
1 - Apigenin
+
+
-
+
-
-
-
-
-
-
-
-
2 - Apigenin-7-O-glycoside
++
+
-
-
-
-
-
-
-
-
-
-
3 - Luteolin-7-O-diglycoside
-
-
+
+
++
++
+
-
-
-
-
-
4 - Luteolin-4`-O-Me (Diosmetin)
-
-
-
-
++
+
-
++
++
-
-
-
5 - Diosmetin-7-O-glycoside
-
-
-
-
t
+
++
-
+
++
-
+
6 - Diosmetin-7-O-diglycoside
-
-
-
-
-
-
+
-
-
+
++
++
7 - Kaempferol-3-O-glycoside
++
++
+
-
-
-
-
-
-
-
-
-
8 - Kaempferol-3-O-diglycoside
-
-
-
-
+
t
+
+
-
t
-
-
9 - Kaempferol-3,7-O-diglycoside
T
-
-
t
+
+
-
++
+
-
+
+
10 - Quercetin (Q)
+
-
t
-
t
t
-
-
-
-
-
-
11 - Q-3-O-glycoside
++
-
+
+
-
-
-
-
-
-
-
-
12 - Q-3-O-rutinoside (Rutin)
++
++
++
++
++
++
++
++
++
++
++
++
13 - Q-3,7-O-diglycoside
-
-
-
-
-
-
+
T
t
+
+
-
14 - Q-3-O-diglycoside-7-O--glycoside
-
t
t
t
-
+
+
+
-
++
++
++
Table 5: The distribution of flavonoids in Galium species collected from different localities of libya. For localities of Galium species.
In addition to the main flavonol compound (12), there are three other major flavonoids in Galium setaceum collected from Wadi Derna identified as apigenin-7-glycoside (2), kaempferol-3-glycoside (7) and quercetin-3-glycoside (11), accompanied by less major amount of apigenin (1) and quercetin (10), whereas kaempferol- 3,7-diglycoside (9) present in trace amount. Galium aparine was represented by three samples collected from various locations of Libya showed resemble flavonoids pattern, but qualitatively different. These samples have a major quercetin-3-rutinoside (12), along with trace amount of quercetin-3-diglycoside-7-glycoside (14). Additionally, sample collected from Gasr Libya contained major amount of kaempferol-3-glycoside (7) with less major amount of apigenin (1) and its 7-glycoside (2). However, G. aparine was collected from Shahat Ruins showed less major amounts of luteolin-7-diglycoside (3), kaempferol-3-glycoside (7) and quercetin-3-glycoside (11), along with trace amount of quercetin (10). On the other hand, apigenin (1), luteolin-7-diglycoside (3) and quercetin-3-glycoside (11) were isolated from G. aparine that collected from Wadi qaam (khoms) in less major amount, but kaempferol-3,7-diglycoside (9) is present in trace value.
With respect to Galium tricornutum, qualitative and quantitative variations of flavonoid profiles characterized by the presence of luteolin-7-diglycoside (3), diosmetin (4) and its 7-glycoside (5) and 7-diglycoside (6) with complete absence of apigenin (1) and its 7-glycoside (2), as well as the 3-glycoside of both kaempferol (7) and quercetin (11). Sample collected from Stwa afforded seven flavonoids including major amount of compounds (3), (4) and (12), along with lesser content of glycosides (8) and (9), while the flavonoids (5) and (10) detected in trace amount. There are two major flavonoids: (3) and (12) in G. tricornutum collected from Lamluda, in addition to less major compounds of (4), (5), (9) and (14) along with trace level of compounds (8) and (10). On the other hand, the major flavonoids isolated from G. tricornutum collected from Almansoura are diosmetin-7-glycoside (5) and quercetin-3-rutinoside (12), along with less major amount of flavone derivatives (3) and (6), as well as flavonol glycosides (8), (13) and (14). Galium verrucosum and G. murale showed a nearly similar pattern of flavonoid compounds, particularly the methylated luteolin (diosmetin) (4), its 7-glycoside (5) and 7-diglycoside (6); with complete absence of the aglycones and simple substituted compounds including apigenin (1) and its 7-glycoside (2); luteolin-7-diglycoside (3); kaempferol-3-glycoside (7); quercetin (10) and its 3-glycoside (11). The flavonoid patterns of G. verrucosum collected from Almansoura characterized by the presence of higher amount of compounds (4), (9) and (12), accompanied by lesser content of compounds (8) and (14), as well as trace amount of flavonoid (13). In addition, sample collected from Wadi Derna contained higher levels of compound (4) and (12), with less major content of compounds (5) and (9) and trace level of flavonoid (13). On the other hand, G. verrucosum collected from Wadi qaam (khoms) has major amounts of flavonoid (5), (12) and (14), along with less major amount of compounds (6) and (13), while compound (8) present in trace level.
The two samples of Galium murale collected from Wadi Elkouf and Sharshara (Tarhuna) contained nearly similar flavonoid patterns with respect to the predominant of diosmetin-7-diglycoside (6), quercetin-3-rutinoside (12) and quercetin-3-diglycoside-7-glycoside (14), along with less major amount of compound (9). However, the quercetin-3,7-diglycoside (13) was isolated only in the first sample that replaced by diosmetin-7-glycoside (5) in the second sample only.
Multivariate analyses: Morphologically similar and taxonomically closer species have similar even same flavonoid profiles. So, they clustered together in the dendrogram. Three groups of species can be separated in the dendrogram (Figure 1). The PCA biplot in (Figure 2) showed that each group is correlated to more than one compound.
Chemotaxonomic significance: This study confirmed that flavonoid content is a useful marker in the taxonomy of Galium. Kaempferol-3,7-O-diglycoside and Quercetin-3-O-rutinoside (Rutin) are found in all species. The isolated and structure elucidation of flavonoid compounds afford the presence of simple and relatively complex flavonoids identified among the five Galium species. The flavonoid complexity appeared in the substitution patterns of flavone and flavonol aglycones with O-glycosylated and O-methylated attachments. Generally, the flavonoid chemistry in all studied Galium species appears to be relatively homogeneous based on the predominance of flavonol glycoside, particularly quercetin-3- rutinoside (12).
The first group (group C) included Galium setaceum and G. aparine, which are characterized by simple O-glycoside of apigenin, kaempferol and quercetin. However, the detection of luteolin-7- diglycoside (3) along with trace amount of complex O-glycosylated quercetin (14) only in Galium aparine and their lacking from G. setaceum indicates the more specialized of the former species relative to the second one. Galium verrucosum and G. murale represent the second group (group B), which was encountered to produce methylated flavone: diosmetin (4) and its glycosidic attachments (5) and (6), along with the presence of complex O-glycosylated kaempferol and quercetin, particularly a C-3 and C-7 positions (9 and 14). In addition, the frequent lacking of simple aglycones of apigenin (1) and quercetin (10), as well as their simple glycosidic forms (2), (3), (7) and (11) indicates that both species are of evolutionary advanced than the two species of first group.
With respect to their classification, Group (A) comprised of Galium verrucosum (S9, S10) and G. tricornutum (S2, S7). Morphologically, these two species are linked together as they share some characters such as mericarp size, petal length, flower diameter size, and petal width. They are clustered together as a result of the same flavonoid profiles (Figure 2). Group (B) included G. murale (S5, S8), G. verrucosum (S11) and G. tricornutum (S3) as well. The former species is characterized by its seed and mericarp shapes. Group (C) included two different species: G. setaceum (S6) and G. aparine (S1, S4, S12). Both species are correlated with petal colour, style length, leaf width, leaf shape, and pedicel length. Remarkably, G. aparine (S1, S4, S12) that collected from distant mountainous and isolated areas do not affected by any flavonoid except for the two which are common to all species. The phytochemical results are relatively congruent with morphological data.
However, qualitative and quantitative variations in the flavonoid profiles among the five investigated Galium species collected from Libya could respond to the environment-dependent variability [24]. Such change may be related to genetic variation amongst individuals that affected by environmental factors. On the other hand, Puff [25] described that variation change in the quality and quantity in the flavonoid profiles in the most taxa of Galium sect. aparinoides is related to change of the environment. Separate publications on the effect of some environmental variables, and genetic variations among the studied Galium are in preparation.
In Conclusion, the flavonoid patterns in the Galium species showed a predominant flavonol glycoside being quercetin-3- rutinoside [26]. However, the other compounds were variable in quality, quantity and in their distribution among the investigated samples. The distribution patterns of the isolated fourteen flavonoid compounds showed the presence of biosynthetic divergence in the glycosidic and methylated attachments that possible to distinguish between two main groups with an intermediate group.
References
- Mabberley DJ. The Plant-Book, Cambridge University Press, Cambridge, New York, Melbourne. 1997.
- Dempster LT, Delprete PG, Smith LB, Klein RB. Rubiáceas. Galium. Flora ilustrada catarinense. Herbário Barbosa Rodrigues, Itajaí. 2004; 273-342.
- Schischkin BK. Flora of the USSR, Delhra Dunn, India: Bishen Singh Mahendra Pal Singh. Koeltz Scientific Books, Koenigstein. 2000.
- Borisov MI, Komissarenko NF. Flavonoids of Galium palustre. Chemistry of Natural Compounds. 1969; 5: 371-375.
- Chen LJ, Bin LL, Shi Jun L, Jun Xing D. Study on the chemical constituents of Galium aparine L. Journal of International Pharmaceutical Research. 2010; 37: 387-389.
- El-Gamal AA, Koichi T, Itokawa H, Halim AF, Amer MM, Saad HEA, et al. Anthraquinones from the polar fractions of Galium sinaicum. Phytochemistry. 1995; 40: 245-251.
- Schildknecht H, Straub F, Scheidel V. Mollugin a new pigment from rhizomes of Galium mollugo. Justus Liebigs Annals of Chemistry. 1976; 8: 1295–1306.
- Borisov MI. Flavonoids of Galium ruthenicum. Chemistry of Natural Compounds. 1974; 10: 662-663.
- Iavarone C, Sen A, Trogolo C, Villa S. Mollugoside, an iridoid glucoside from Galium mollugo. Phytochemistry. 1983; 22: 175-178.
- Zhao CC, Shao JH, Li X, Kang XD, Zhang YW, Meng DL, et al. Flavonoids from Galium verum L. Journal of Asian Natural Products Research. 2008. 10: 611-615.
- Zhao CC, Shao JH, Cao DD, Zhang YW, Li X. Chemical constituents of Galium verum L. China Journal of Chinese Materia Medica. 2009; 34: 2761-2764.
- Corrigan D, Timoney R, Donnelly D. Iridoids and alkanes in twelve species of Galium and Asperula. Phytochemistry. 1978; 17: 1131-1133.
- Uesato S, Miyauchi M, Itoh H, Inouye H. Biosynthesis of iridoid glucosides in Galium mollugo, Galium spurium var. echinospermon and Deutzia crenata. Intermediacy of deoxyloganic acid, loganin and iridodial glucoside. Phytochemistry. 1986; 25: 2515–2521.
- Baytop T. Therapy with Medicinal Plants in Turkey: Past and Present. Istanbul University Publications. 1984.
- Grzybek J, Wongpanich V, Mata-Greenwood E, Angerhofer CK, Pezzuto JM, Cordell GA. Biological evaluation of selected plants from Poland. International Journal of Pharmacognosy. 1997; 35: 1-5.
- Boulos L, Barakat HN, Hegazy AK. Endemic Flora of the Middle East and North Africa. Reviews in Ecology: Desert Conservation and Development. 1997; 229-260.
- Brullo S, Furnari F. La vegetation del Gebel el-Akhdar (Cirenaica settentrionale). Boll Acc Gioenia Sci Nat. 1994; 27: 197-412.
- Le Houérou HN. Biogeography of the arid steppeland north of the Sahara. Reviews in Ecology: Desert Conservation and Development. Cairo: Metropole, Egypt. 1997; 207-228.
- Feng Y, Lei J, Xu X, Pan R. Composition and characteristics of Libyan flora. Archives of Biological Sciences, Belgrade. 2013; 65: 651-657.
- Soltis D, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sinauer, Sunderland. 2005.
- Mabry TJ, Markham KR, Thomas MB, Mabry TJ, Markham KR, Thomas MB. The Systematic Identification of Flavonoids. Springer-Verlag and New York. 1970.
- Hammer ø, Harper DAT, Ryan PD. PAST: Palentological Statistics software package for education and data analysis. Palaentologia Electronica. 2001; 4: 9.
- Kovach WL. A Multivariate Statistical Package for Windows, Manual. Kovach Computing Services, Pentraeth, Wales, UK. 1999.
- Ehrendorfer F, Schönbeck-Temesy E, Puff C, Rechinger W. Rubiaceae Rechinger, Flora Iranica. Verlag des Naturhistorischen Museums Wien, Vienna, Austria. 2005.
- Harborne JB. Comparative Biochemistry of the Flavonoids. Academic Press, London and New York. 1967.
- Puff C. Leaf flavonoids of Galium Sect. Aparinoides (Rubiaceae). Plant Systematics and Evolution. 2001; 124: 57-66.