
Short Communication
Austin J Plant Biol. 2017; 3(1): 1017.
First Report of Bhendi Yellow Vein Mosaic Virus in Tabernaemontana Divaricata and Albizia Saman in India
Roy B, Sultana S and Sherpa AR*
Department of Botany, West Bengal State University, India
*Corresponding author: Ang Rinzing Sherpa, Department of Botany, West Bengal State University, Kolkata, India
Received: May 17, 2017; Accepted: June 21, 2017; Published: June 29, 2017
Abstract
During survey from March 2014 to October 2015, in Barasat, West Bengal, India, incidence of symptoms suggestive of virus infection was observed in Tabernaemontana divaricata (family –Apocynaceae) and Albizia saman (Fabaceae). Infected plants were showing typical symptoms of Begomovirus infection, including leaf curling, leaf yellowing and stunted growth. The occurrences of geminivirus were confirmed by symptomatology, southern blot analysis, Polymerase Chain Reaction (PCR) and nucleotide analysis of the part of genome. Sequence analysis showed that the virus from both the plants share 93-99% identity with the sequences of other BYVMV isolates. The sequence from T. divaricata and A. saman share 99% identity with each other at the nucleotide level showing close phylogenetic relationship. Host range study shows that viruses from both the plants can transfer to chilli plant through mechanically. This is the first molecular evidence of Bhendi yellow vein mosaic virus infecting T. divaricata and A. saman in West Bengal, India.
Keywords: Begomovirus; PCR; Southern blot; Bhendi yellow vein mosaic virus; Geminivirus; Detection
Introduction
Viruses of the genus Begomovirus are transmitted by the whitefly Bemisia tabaci and are the most numerous and economically destructive viruses among the geminiviruses. They comprise the largest numbers of species (288 of 325 total species) in the family Geminiviridae [1]. During the last 30 years, begomoviruses have emerged as important viral pathogens in the field that infect a broad variety of food, fiber, and ornamental crops and cause significant losses worldwide. Begomoviruses have been sub-divided into two types with either a bipartite or monopartite genome [1,2].
Bhendi Yellow Vein Mosaic Virus (BYVMV) is a member of whitefly-transmitted geminiviruses of the genus Begomovirus. It is a single stranded DNA virus and is important pathogen of a wide range of crop ecosystems. The majority of begomoviruses have a genome comprising two similar sized DNA components (DNA A and DNA B). The DNA A component encodes a Replication-associated protein (Rep) that is essential for viral DNA replication, a Replication Enhancer protein (REn), the Coat Protein (CP) and a Transcription Activator Protein (TrAP) that controls late gene expression. The DNA B component encodes a Nuclear Shuttle Protein (NSP) and a Movement Protein (MP), both of which are essential for systemic infection of plants [3,4]. In contrast, some begomoviruses have only a single DNA A genomic component, such as isolates of Cotton Leaf Curl Virus (CLCuV), Tomato Yellow Leaf Curl Virus (TYLCV), Ageratum Yellow Vein Virus (AYVV), and Tomato Leaf Curl Virus (TLCV) [5-9]. In 1924, Bhendi yellow vein mosaic virus was first reported in okra plants from India and Sri Lanka. Okra or Bhendi plants infected by BYVMV shows symptoms of vein clearing and yellowing. Leaves and fruits are reduced in size and causes significant decrease in the production of the vegetable. Up to 96% loss in yield has been reported [10].
In the time survey from March 2014 to October 2015, in Barasat, West Bengal, India, highly incidence of symptoms suggestive of virus infection was observed in Tabernaemontana divaricata and Albizia saman. Infected plants were showing typical symptoms of Begomovirus infection, including leaf curling, leaf yellowing and stunted growth. T. divaricata belongs to the family Apocynaceae, commonly called Togor, Dudhphul in Bangladesh, pinwheel flower, crape jasmine in India, is an evergreen shrub have been reported to be great sources of various alkaloids. It has been used in traditional medicine as components of rejuvenating and neurotonic remedies [11]. The crude ethanol extract of T. divaricata (L) might possess antinociceptive activity [12]. On the other hand A. saman is a species of flowering tree in the family Fabaceae and reported to possess multiple medicinal effects. It has evident from the study that A. saman has allelopathic effect [13]. The leaves of A. saman display antimicrobial activity [14], and its secondary compounds could be used for therapeutic purposes.
For the identification of the Begomovirus the symptomatic leaf of T. divaricata (family –Apocynaceae) and A. saman (Fabaceae) were collected from the Barasat, West Bengal in India and stored in -80 °C for future investigation (Figure-1a and 1b).

Figure 1: (a) Symptoms of BYVMV in Albizia saman showing chlorosis
and yellow mosaic on leaf. (B) Symptoms of BYVMV in Tabernaemontana
divaricata showing leaf curling, chlorosis and yellow mosaic on leaf.
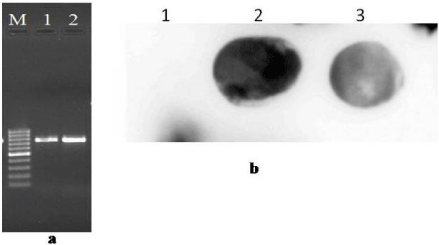
Figure 2: (a) PCR amplification of BYVMV containing AV1, AC3, AC2 gene
fragment of about, 760 bp from nucleic acid extracted from BYVMV –infected
leaves of A. saman (lane -1), T. divaricata (Lane 2) and Lane M: 100 bp
DNA Marker (Black Bio). (b) Southern hybridization detection of BYVMV by
using biotin probe, lane 2- A. saman and lane 3-T. divaricata were positive in
southern hybridization (lane 1 were found negative).
For the detection of begomoviruses, two detection techniques were used: 1) PCR amplification of virus by using indigenously designed geminivirus specific primer pair and 2) using Southern blot analysis using biotin-labeled probes specific for geminivirus.
Total DNA was extracted from infected leaves using our new modified CTAB method [15] and tested for the presence of geminiviruses by PCR using indigenously designed geminiviruses specific degenerate primer pair [16]. The primers were designed by multiple alignment of the DNA-A of geminiviruses sequences available in GenBank database. The PCR was performed under following condition: initial denaturation 95°C for 5 min, following by 35 cycles of 94°C for 30 sec, 48°C for 45 sec, 72°C for 1 min, and final extension at 72° C for 7 min. The expected fragment size of the amplicon was about 760 nt. The PCR products were eluted from 1% agarose using the gel extraction kit (XcelGen- Xcelris Genomics) and sent for sequencing.
For the Southern blot analysis we designed a biotin labeled probe and used for the detection of the geminiviruses from the total sap of the infected plant samples. Briefly, about 5 μl of freshly prepared sap was blotted on the nitro cellulose membrane and air dried the membrane properly. After that the membrane was UV-cross linked for 30 min under UV-Cross Linker (GeNeiTM, India). Prehybridization, hybridization and washing of membrane were done according to the southern blot analysis protocol using biotin-labeled probes [17].
The mechanical transmission of both the virus obtains from T. divaricata and A. saman were conducted with sap inoculation. Briefly, the infected T. divaricata and A. saman leaves were crushed in 0.1 M potassium phosphate buffer, pH 7.0 (1:2, w/v) containing 0.15 % sodium sulphite. The sap from infected plants leafs were used to inoculate the healthy chilli plants (Capsicum annuum), with the help of carborundum powder [18]. Five different plants were selected for transmission test. Initially, the young leaves of test plants were dusted with carborundum powder, and then a cotton-wrapped stick was soaked in the sap inoculums. The stick was rubbed on the leaves gently where carborundum powder was previously applied. Inoculated plants were kept in a net house for symptom development. The visible symptoms appeared after transmission experiment confirmed the success mechanical transmission of the viruses.
Positive PCR reaction from T. divaricata and A. saman indicates the Begomovirus infection in the plants which amplified the parts of AV1, AC3 and AC2 genes fragment of approx ~760 bp in length from symptomatic plants, but were absence in symptomless samples (Figure 2a). PCR products were suitably cloned into pGEM-T vector and sequenced, and submitted in the GenBank database as accession number; LC010948 and LC010949 respectively.
In Southern hybridization technique all samples from symptomatic plants hybridized with the probe, whereas samples extracted from non – symptomatic plants did not show positive results (Figure 2b). Hybridization of Begomovirus probe with the DNA samples on the nitrocellulose membrane indicates that these probes can also be used for the detection of begomoviruses. The strong signal showed that the virus titer in T. divaricata and A. saman were high.
Sequence of PCR product were analyzed with available Begomovirus DNA-A sequences obtained from the GenBank database using Multalin, MUSCLE, BLASTn, and pair wise identity scores were calculated using SDTv1.2 (Sequences Demarcation Tool version 1.2). SDTv1.2 result analysis of the virus from T. divaricata and A. saman showed that the both virus shares 78 % identity with that of a Mesta yellow vein mosaic virus isolate from Haringhata (Acc. No.- FJ159263), 73% with Tomato leaf curl virus isolate from Barasat (Coccinia grandis, unpublished) (Acc. No.- LC010942), 99% with Bhendi yellow vein mosaic virus isolate from Haryana and Kalyani respectively (Acc. No.- FN645923 and KJ462082) and shares identity with each other 99% at nucleotide level (Figure 3).
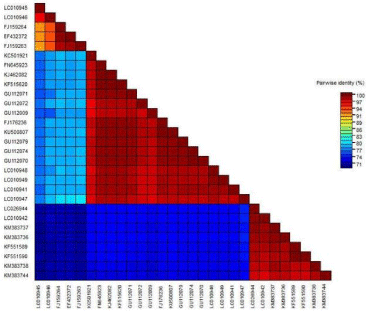
Figure 3: Colour coded matrix of pairwise similarity scores of BYVMV with
other begomoviruses by SDT (Sequences Demarcation Tool version 1.2).
Phylogenetic tree was constructed using Vector NTI, BioEdit and Neighbor- joining analysis with Phylip programs. Banana bunchy top virus acc. No: EU140341 was used as an outgroup and marked with solid square. Our sequences were marked with solid triangle (Figure 4). During phylogenetic analysis of PCR product comprising part of AV1, AC3 and AC2 genes of the begomovireses under study (Acc. No.- LC010948 and LC010949) together with isolates of BYVMV, ToLCV and MYVMV formed three independent clusters. All other BYVMV isolates (Acc. No.- EU140341, LC010947, KC501921, LC010941, KF515620, KJ462082, FN645923, GU112072, GU112071, GU112009, GU112079, GU112074, GU112070, KU500807 and FJ176236) and our isolates (Acc. No.- LC010948 and LC010949) clustered distinctly together in cluster I, while isolates of ToLCV (Acc. No.- KF551590, KF551589, KM383744, KM383738, KM383737, KM383736, LC026944 and LC010942) clustered together in cluster II. MYVMV isolates (Acc. No.- LC010945, LC010946, FJ159263, FJ159264 and EF532372) clustered separately into cluster III.
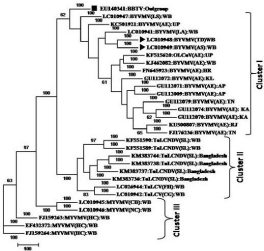
Figure 4: Evolutionary relationships and similarities of BYVMV with other
begomoviruses (TolCNDV: Tomato leaf curl New Delhi virus, MYVMV: mesa
yellow mosaic virus and OLCuV: Okra leaf curl virus.
For host range and transmission study, the viruses were inoculated into the healthy chilli plant mechanically. The characteristic symptoms were observed on C. annuum, 10-15 days post inoculation (dpi) after mechanical inoculation of the virus. The virus was found to be transmitted by mechanical transmission at transmission efficiency of 100 %. A positive PCR amplification of 760 bp sized products was obtained with the DNA-A specific primer sets. Sequencing results confirmed the virus infection.
Our results also confirms that the viruses in study infects T. divaricata and A. saman in Barasat, India indicating potential threat to other cultivated crops and both the hosts acting as a virus reservoir. However, there is a need for more detailed study about the viruses as it is broadening its host range. Many geminiviruses are already reported as emerging and re-emerging in the recent years and infecting different host and threatening the economically important crops which are susceptible for geminiviruses and therefore it is essential to study the survey and detection of the viruses in other crops, spread of the disease and characterize the viruses in detail at molecular level. The study of molecular interaction between the host and vectors of the viruses, explorations into the ecology of these viruses, including interactions with other viruses are also important areas to focus in future research. To the best of our knowledge this is the first record of T. divaricata and A. saman as a natural host of BYVMV.
Acknowledgement
Authors would like to thank Department of Science and Technology, New Delhi for SERB Project Grant No. SR/FT/LS- 165/2012 and financial assistant from University Grant Commission (UGC), New Delhi to Mr. Roy B, Senior Research Fellow (Rajiv Gandhi National Fellowship) in the form of fellowship is greatly acknowledged.
References
- Brown JK, Fauquet CM, Briddon RW, Zerbini FM, Moriones E, Navas- Castillo J. Geminiviridae: Virus Taxonomy: Classification and Nomenclature of Viruses - Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier London. 2012; 351-373.
- Seal SE, van den Bosch F, Jeger MJ. Factors influencing Begomovirus evolution and their increasing global significance: implications for sustainable control. Critical Reviews in Plant Sciences. 2006; 25: 23-46.
- Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci. 1999; 18: 71–106.
- Gafni Y, Epel BL. The role of host and viral proteins in intra- and inter- cellular trafficking of geminiviruses. Physiol Mol Plant Pathol. 2002; 60: 231–241.
- Kheyr-Pour A, Bendahmane M, Matzeit V, Accoto GPM, Crespi S, Gronenborn B. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res. 1991; 19: 6763–6769.
- Navot N, Pichersky E, Zeidan M, Zamir D, Czosnek H. Tomato yellow leaf curl virus: A whitefly-transmitted geminivirus with a single genomic component. Virology. 1991; 185: 151–161.
- Dry IB, Rigden JE, Krake LR, Mullineaux PM, Rezaian MA. Nucleotide sequence and genome organization of tomato leaf curl geminivirus. J Gen Virol. 1993; 74: 147–151.
- Tan PHN, Wong SM, Wu M, Bedford ID, Saunders K, Stanley J. Genome organization of Ageratum yellow vein virus, a monopartite whitefly-transmitted geminivirus isolated from a common weed. J Gen Virol. 1995; 76: 2915–2922.
- Briddon RW, Markham PG. Cotton leaf curl virus disease. Virus Research. 2000; 71: 151–159.
- Pun KB, Doraiswamy S. Effect of age of okra plants on susceptibility to Okra yellow vein mosaic virus. Indian J Virol. 1999; 15: 57–58.
- Ingkaninan K, Temkitthawon P, Chuenchom K, Yuyaem T, Thongnoi W. Screening for acetylcholinesterase inhibitory activity in plants used in Thai traditional rejuvenating and neurotonic remedies. J Ethnopharmacol. 2003; 89: 261-264.
- Sharker SM, Chakma S, Rahman AA. Phytochemical and antinociceptive study of Leaves of Tabernaemontana divaricata (L) J of Med Pl Res. 2011; 5: 245-247.
- Ghosh R, Paul S, Ghosh SK, Roy A. An improved method of DNA isolation suitable for PCR based detection of begomoviruses from jute and other mucilaginous plants. J Virol Methods. 2009; 159: 34-39.
- Singh A, Tripathi P, Srivastava A, Marzia Ali S, Rekhi L. Antibacterial activity of six indigenous Indian plants: Acacia nilotica (Fabaceae), Albizia saman (Fabaceae), Azadirachta indica (Meliaceae), Carica papaya (Caricaceae), Cymbopogon citratus (Poaceae) and Mangifera indica (Anacardiaceae). Afr J Biotechnol. 2016; 15: 666-669.
- Roy B, Sherpa AR. Extraction of High Quality DNA from Mucilaginous Plants with a New Improved Method, Suitable for Detection of Geminiviruses and Downstream Applications. International Journal of Science and Research. 2017; 6: 121-124.
- Roy B, Chakraborty B, Mitra A, Sultana S, Sherpa AR. Natural occurrence of Bhendi yellow vein mosaic virus on Litsea spp. in India. New Dis Rep. 2015; 31: 7.
- Weigel K, Pohl JO, Wege C, Jeske H. A population genetics perspective on geminivirus infection. J Virol. 2015; 89: 11926 –11934.
- Chakraborty S, Pandey AK, Banerjee MK, Kalloo G, Fauquet CM. Tomato leaf curl Gujarat virus, a new Begomovirus species causing severe leaf curl disease of tomato in Varanasi, India. Phytopathology. 2003; 93: 1485-1495.