
Review Article
Austin J Plant Biol. 2021; 7(1): 1028.
microRNA Regulation of Nodule Zone-Specific Gene Expression in Soybean
Ehsanul Haque ME*
South Dakota State University, Brookings, South Dakota, USA
*Corresponding author: Md. Ehsanul Haque, South Dakota State University, Brookings, South Dakota, USA
Received: June 01, 2021; Accepted: July 06, 2021; Published: July 13, 2021
Abstract
Nitrogen is a paramount important essential element for all living organisms. It has been found to be a crucial structural component of proteins, nucleic acids, enzymes and other cellular constituents, which are inevitable for all forms of life. In the atmosphere, the percentage of nitrogen is very high (N2, 78%) compared to other inorganic gases. However, most organisms have practically no direct access to this nitrogen. While plants cannot directly uptake nitrogen from atmosphere, they are capable of assimilating other forms of nitrogen for example ammonium (NH4+) and nitrate (NO3-). For agricultural crop production, artificial fixation of nitrogen is heavily utilized and it is an expensive process that requires high temperatures (at least 400°C) and pressures (around 200atm). It has been conspicuously demonstrated that indiscriminate use of fertilizer hampers soil physical, chemical and micro biological properties and a potential risk to environment e.g. water quality. Besides, chemically manufactured fertilizers are depleted from soils in various ways, for instance; denitrifying bacteria, volatilization, and leaching. Consequently, it results relatively poor availability of nitrogen to get into plants. On the flipside, only 1-2% of the nitrogen fixation in the world occurs through the natural process of lightening. Notably, microbial fixation is well characterized in diazotrophs for example: Rhizobia and Frankia, and blue-green algae. Against the backdrop, we are accentuated on an environmentally friendly and the most sustainable approach to increase productivity for legume and non-legume crops. Till today, the term Biological Nitrogen Fixation (BNF) has received much attention as a sustainable alternative; this process facilitates atmospheric nitrogen to convert into ammonia by rhizobia in specialized plan organs termed “root nodules”. This review article seeks to better understand plant mechanisms involved in the development of root nodules in soybean.
Keywords: nitrogen; Biological Nitrogen Fixation; Soybean
Introduction
Soybean (Glycine max) is one of the most important oil crops and a source of animal feed protein in the world. It has a salient feature to fix atmospheric nitrogen through symbioses with compatible rhizobia that yields to determinate type nodule. Biological nitrogen fixation in soybean nodules reduces the use of chemical nitrogen fertilizers resulting in cost-savings to producers and minimizes environmental damage due to nitrogen run-off. A better understanding of how nodules form and function is important for selection or generation of soybean genotypes with better nitrogen fixation capacity. Soybean nodules originate from root cortex via de novo cell differentiation. Consequently, two major nodule development zones are formed for instance; the Nodule Primordium (Npr) in the middle and it is encircled by Nodule Parenchyma (Npa). At later time point, the Npr gives rise to N-fixation zone and the Npa holds vascular bundles. It is not clear what early signaling pathways driving the conspicuous development of the nodule zones. My research is aimed at filling this knowledge gap by illustrating the molecular signatures that paves the way to cellular differentiation in root nodule development in soybean. Based on initial evidence obtained by the Subramanian lab, we hypothesize that microRNAs (miRNAs) play important regulatory roles in spatio-temporal expression of their target genes during nodule developmental in soybean. For instance, the regulation of auxin sensitivity by miR160 has been found to be crucial for formation of nodule primordia and vasculature in the parenchyma [1]. Against this backdrop, this review article focused on nuclear and cytoplasmic transcriptome as well as miRNA profiles of parenchyma and primordial tissues, determine the relative abundance, and differentially expressed mRNAs and regulatory role of miRNAs in cell differentiation and nodule development.
Root Nodule a Sustainable Alternative to Fix Atmospheric Nitrogen
Atmospheric nitrogen percentage is very high (N2, 78%) compared to other inorganic gases. However, most of the organisms have practically no direct access to this nitrogen. Nevertheless, plants cannot directly uptake nitrogen from atmosphere but they are capable of assimilating only very specific forms of nitrogen, for example ammonium (NH4+) and nitrate (NO3-) [2]. Virtually, nitrogen has been found to be a crucial structural component of proteins, nucleic acids, enzymes, and other cellular constituents, which are inevitable for all forms of life. For agricultural crop production, artificial fixation of nitrogen is heavily utilized. An expensive process requires high temperatures (approx. 400°C) and pressures (approx. 200atm). It has been conspicuously demonstrated that indiscriminate use of N fertilizer hampers the diversity of the bacterial community and decreases soil C and N concentrations. Notably, it has been demonstrated as a potential risk to environment e.g. water quality [2]. Besides, chemically manufactured fertilizers are depleted from soils in various ways, for instance; denitrifying bacteria, volatilization, and leaching [3]. Consequently, it results relatively poor availability of nitrogen to get into plants. On the flipside, over 90% of the nitrogen fixation in the world occurs through the natural process of lightening and microorganisms. Furthermore, microbial fixation is well characterized in diazotrophs for example; Rhizobia and Frankia, and blue-green algae [4]. It has been demonstrated that Bradyrhizobium strains substantially escalated soybean grain yield, and protein content up to 57% and 26%, respectively. Against the backdrop, we are accentuated on an environmentally friendly and a sustainable approach to increase the productivity for legume and non-legume crops. Literature mining depicted that biological nitrogen fixation in soybean nodules reduces the use of chemical nitrogen fertilizers resulting in cost-savings to producers and minimizes environmental damage due to nitrogen run-off.
Rhizobia Infection Leads to the Root Nodule Development
In the natural environment, plants are continuously confronted with pathogenic and symbiotic microbes. Symbioses involves mutual exchange of diffusible signal molecules, first endophytic bacteria (rhizobia) are attracted by the plant root exudates flavonoids, which are perceived and triggered the bacterial nodulation (nod) genes. Consequently, the bacteria synthesize specific lipochitooligosaccharides, called nodulation (Nod) factors. This signal is perceived by the LysM receptor like kinase of host plant, it induces the root hair curling, and bacteria get access into the host epidermis through Infection Threads (ITs) and initiate cell division within the root cortex, leading to the progression of the root nodule meristem. In later stages of the interaction, bacteria are released from the infection threads into the plant cells, surrounded by membrane of plant origin. These bacteria multiply within the host cells and differentiate into the nitrogen fixing bacteroids [5]. Till now, integration of genetic and genomic approaches has revealed twenty-six genes to be involved in nodule development of Medicago truncatuala and Lotus japonicum. In addition, deep sequencing of the Medicago truncatula root transcriptome has uncovered thousands of genes to be induced during Nod factor signaling and its resulting ethylene (ET) biosynthesis throughout the multiple development stages of indeterminate nodule. Albeit the molecular mechanism of such regulation is not well understood. There has been a large-scale transcriptome analysis of B. japonicum-inoculated and mock-inoculated soybean root hairs. It has showed that a total of 1,973 soybean genes differentially expressed during root hair infection, particularly NFR5 and NIN genes. Nevertheless, the signaling mechanisms directing the cellular differentiation of nodule are not known.
Soybean Root Nodule Organogenesis
Soybean (Glycine max) has a genome size of 1.1 to1.5 Gb, it is partially diploidized tetraploid. It is one of the most important oil crops and a source of animal feed protein in the world (soybase.org/ sb_about.php). It has a salient feature to fix atmospheric nitrogen through symbioses with compatible rhizobia that yields to determinate type nodule [5]. Notwithstanding of the economic and environmental importance, there has been very few studies about Quantitative Trait Loci (QTL) that controlling BNF traits, for instance nodule number, ration of nodule dry weight with nodule number, and Shoot Dry Weight (SDW). It has been reported via composite interval mapping that approximately six QTLs bears very small effect on BNF traits. Besides, it has been demonstrated in earlier studies that nodules originate from root cortex via de novo cell differentiation into two different cell types, parenchymal and primordium [6,7]. In addition, early nodulin genes in legume for instance; Enod 40 gene reported to be expressed in root pericycle during the rhizobia infection and later it occupied in the dividing cortical cells [8]. Among the two major nodule development zones, the Nodule Primordium (Npr) in the middle, which is encircled by Nodule Parenchyma (Npa). At later time point, the Npr gives rise to N-fixation zone and the Npa holds vascular bundles. Lately, a β-expansin gene, GmEXPB2 fused with GUS reporter gene, which was observed to be preferentially expressed in nodule vascular trace and nodule vascular bundles. It indicated that GmEXPB2 might be crucial for nodule organogenesis. Over expression of GmEXPB2 contrast to suppressed GmEXPB2 transgenic lines found to be escalated nodule number, nodule mass and nitrogenase activity. It further suggested that GmEXPB2 might have influenced over root architecture, nodule formation and development, and profoundly yielding to biological N2 fixation. Even though, it is not clear what early signaling pathways driving the conspicuous development of the nodule zones. Against the backdrop, to understand the regulation of auxin sensitivity by miR160, which is, believed to be crucial for the formation of nodule primordia [1] (Figure 1).
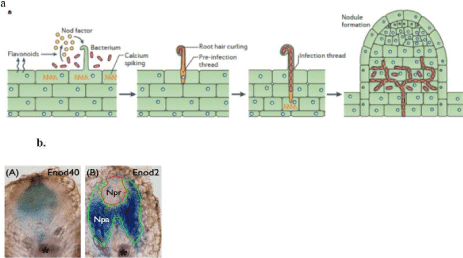
Figure 1: a) Illustrating the progression of root nodule development through Rhizobial bacterial infection in the plant root leading to the determinate nodule.
b) Nodule development zones; A) Nodule primordial zone (Enod 40 gene) in the middle B) Surrounding parenchyma (Enod 2 gene), differentiated from cortex
(collected from Sen Subramanian lab).
Regulatory Small RNAs Biogenesis and its Molecular Functions
Regulatory small RNAs are ranged between 20 to 24 nucleotides, which are ubiquitous elements of endogenous plant transcriptomics, a common response to exogenous viral infections and introduced double-stranded RNA [9]. Three core enzymes families, for instance; RNAdependent RNA polymerase (RDR), Dicer like (DCL), and Argonaute (AGO) proteins paves the way of small RNA biogenesis and function in plants. Firstly, ribonuclease type III or DICERLIKE1 involves in the yield of a fold-back precursor RNA or primary miRNA (primiRNA) transcripts using an RNA templates in the nuclei. Later, the resulting miRNA-miRNA duplex, which is originated in nucleus, then translocated into cytoplasm. The guided miRNA molecule is incorporated into ARGONAUTE (AGO) to form an active RISC complex to specific target RNAs that are complementary to the miRNA, and this process eventually follows up mRNA cleavage, represses the translation of the mRNAs or Chromatin modification. This phenomenon accentuated as an inhibition or silencing of the gene expression, which play a crucial role in the developmental process in plant and animal [9,10] (Figure 2 and Figure 3).
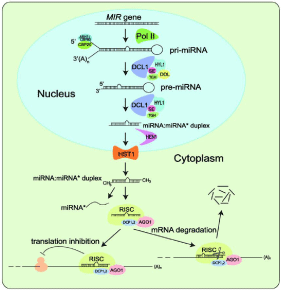
Figure 2: Regulation of gene expression events via RISC complex (modified
from https://www.google.com/?gws_rd=ssl#q=mirna+picture+in+plants
accessed on February 7, 2016).
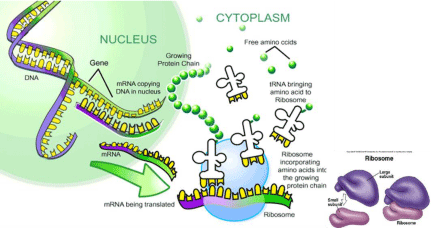
Figure 3: Gene expression events occurring in typical plant cell (modified https://www.google.com/search?q=transcription+and+translation accessed on February
7, 2016).
It has been found in several studies that most plant miRNAs are non-coding RNA and small 21-24 nucleotide long [1]. It requires DCL1-clade DCL for their biogenesis and AGO1-clade AGO for their function [12,13].
In rice (Oryza sativa), DCL3 has been reported in the biogenesis of 24nt long miRNA that incorporated in AGO4 to regulate the target gene expression primarily through mRNA cleavage [12]. Argonaute Proteins (AGO) form RNA Inducing Silencing Complexes (RISC) with small RNAs, which is known as post-transcriptional gene silencing. It has typically four domains, for instance: N-terminal, PAZ, MID and PIWI domains. The MID-PIWI lobes are belongs to the C-terminus. It has been studied that MID-domains contains the specificity loop to recognize and bind to the 5’-phosphate of smRNAs. The PIWI domains contained the catalytic active site D-E-D-H/D. PAZ domain anchored the 2-nt overhang at the 3’ end of miRNAs. The N-terminal domain involved in the separation of miRNA-miRNA duplex and the slicer activity of the mRNA [14]. There has been an expansion and duplications of AGO family members during plant evolution [15]. The functional diversification of AGOs is indicating sRNA directed regulatory pathways. The binding preference of AGO and sRNA is mainly assigned by the sequence of sRNA. In Arabidopsis, 10 AGO have been extensively studied. It has been demonstrated that AtAGO10 like AtAGO1, it recognized distinct structural features in miR165/miR166 duplex than involved by AtGO1. AtAGO10 found to regulate shoot apical meristem by decoying miR165/miR166 and subsequent repression of Homeodomain-Leucinezipper (HD-ZIP) gene expression [16]. Notably, 22 AGO proteins have been reported in Soybean (Glycine max). It has been found that genome duplication in Soybean resulted such a proliferation of AGOs. For example: Its genome encodes two copies of AGO1, AGO2, AGO5, AGO4/9, AGO6 and AGO7 [17]. However, the molecular function of the plant AGO genes yet not very clear.
Several miRNA families are conserved across the vast evolutionary distances from flowering plants to mosses [11]. It has been observed in another study that miRNA, and its target pairing found to be stable for prolonged periods of plant evolution.
On the flip side, another group demonstrated that conserved plant miRNAs and their targets are to somehow flexible. For instance, miR159 is a highly conserved miRNA that targets not only a subset of MYB mRNAs but also observed to target a non-MYB mRNA, SGN-U567133 [18]. A mutant tomato transgenic line (miR159- resistant line) showed higher level of the SGN-U567133 transcript and exhibited defects in leaf and flower development. This result suggests that miR159 involves in a post-transcriptional regulation. Additionally, it is found to be crucial for the normal tomato development.
Recently, the identification of miRNAs in the regulation of photoperiodic pathways in soybean have been reported through high throughput sequencing and qRT-PCR. Six libraries were constructed using Illumina Solexa, for instance; 0, 8, and 16 h under short day treatment, similar time points considered for the long the long day treatment. A total of 163 miRNAs families were reported which covered 318 plant miRNAs, and unclassified 81 novel predicted miRNAs. As expected, significant differences in abundance between short day and long day treatment was observed. These findings provided evidence of miRNA in the regulation of flowering time that ultimately affects the seed yield and quality of soybean.
The complex regulatory network of miRNA-mRNA interactions during viral infection has been revealed via small RNA seq (sRNA), degradome seq, and genome-wide transcriptome analysis.
There has been a total of 253 soybean miRNAs found to be two-folds abundance compared with mock-inoculated control demonstrated through sRNA seq analysis. Among them 105 miRNAs were identified as potential targets of 125 transcripts that has been validated by degradome seq analyses. In addition, 2679 genes were detected via genome wide transcriptomic analysis. These genes have been differentially expressed during infection of soybean mosaic virus and among them 71 genes projected to induce in defense response. These findings suggested the regulatory role miRNA that governed the target gene expression during viral infection.
Furthermore, the regulatory role of microRNAs (miRNAs) during Soybean-Bradyrhizobium japonicum mutualistic association was studied first by Subramanian et al. [19]. They sequenced approximately 350000 small RNAs of soybean root sample, which were inoculated with B. japonicum. It helps to detect 20 conserved miRNAs loci based on the similarity to miRNAs in another plant species. In addition, 35 novel miRNAs were identified based on potential hairpin forming precursors in Soybean EST as well as shotgun genomic sequences [19]. These findings advocated the potential role of miRNAs in the regulation of legume rhizobium symbiosis.
In another study, Turner et al have identified 120 hairpinforming precursor genes in soybean. In addition, they reported three novel miRNAs for instance; miR160, miR164 and miR393 found to be involved in auxin signaling [20]. Moreover, the plant hormone auxin is thought to have a pivotal role in nodule organogenesis in determinate and indeterminate type of nodule. It indicates a redundancy and diversity of miRNAs family members that governs the formation of root nodule.
It has been illustrated that auxin receptor gene family hushed by overexpressed microRNA393. These plant roots found to be hypersensitive to auxin and yielded normal nodule. This observation advocated that only minimal/reduced auxin signaling is required for determinate nodule development. Likewise, overexpressed microRNA160 hushed a set of repressor auxin response transcription factor. These plant roots were hypersensitive to auxin and observed not to be reluctant in epidermal responses to rhizobia. Notably, it yielded to lower sized nodule primordium [1]. This observation indicated that auxin hypersensitivity inhibits nodule organogenesis Organ specific expression of profile of miRNA and the potential targets were also studied. Two genes (Glyma10g10240 and Glyma17g05920) which were the target of miR169 but detected to be highly expressed in soybean nodule. Likewise, three potential targets of gma-new-miR13587 demonstrated to be highly expressed in the nodules than in the roots. As expected, gma-newmiR13587 found to be poorly expressed in the nodules than in the roots [20].
There was an inverse expression pattern observed in between roots and nodules.
Li et al., studied the transgene expression of three novel miRNAs namely, miR482, miR1512, and miR1515 in Soybean. They noticed a significant increase of nodule numbers while root length and later root density were normal in all tested miRNA lines. As expected, there were differential expression of these miRNAs in supernodulating and nonnodulating soybean mutants. They reported that 6 novel miRNAs decoyed 22 predicted target genes. And it was estimated via real time polymerase chain reaction and qRT-PCR [21]. It advocates that miRNAs have the signatory roles in soybean nodule development.
Sequencing of small RNAs and Parallel Analysis of RNA Ends (PARE) libraries revealed to identify 284 nodule miRNAs, more than 500 target genes, and including 178 novel soybean miRNAs. It has been reported that ENOD93 only found to be expressed in nodule tissue not in other plant parts of Soybean. Ectopic expression of miR393j-3p and RNAi silencing approach to ENOD93 expression showed a significant reduction in nodule formation [22].
Therefore, this study showed a list of miRNAs and their potential target of nodulation genes.
In the model legume (Medicago truncatula), 25 conserved miRNA families and 100 novel miRNA reads were detected by highthroughput sequencing. The expression of MtHAP2-1 (encodes a CCAAT binding transcription factor) to meristematic zones was restricted by miR169a which is found to be critical for the development of indeterminate type of nodule [23]. In another study, HDZIPIII transcripts were inhibited by overexpression of miR166, it dropped the number of symbiotic nodule and lateral root [24]. To get insights into key genes of nodule zones, transcript profiles of specific cells/tissues were investigated at different time points from indeterminate nodules of M. truncatula using laser capture micro dissection. It has been demonstrated from the comprehensive gene expression map that selected genes enriched in different cell/tissue types. These findings indicated that organ specific gene expression could be controlled by the presence or absence of miRNAs.
Recently, Agrobacterium rhizogenes mediated hairy root transformation has been applied as tool for exploring cell type specific gene expression in tomato. Cell type or tissue specific promoter introduced into INTACT and TRAP constructs via gateway cloning technology to develop binary vectors. INTACT method used to capture biotin tagged nuceli from specific cell types and TRAP method used for profiling of mRNAs or foot printing of individual ribosomes. TRAP methodology is not required tissue fixation or single cell suspension. It has been successfully used to date in organisms ranging from D. melanogaster to mice and human cultured cells. Multiple ribosomes or Polyribosomes (polysomes) are engaged in translation on a single mRNA. To evaluate the translation state of an mRNA, ribosomal subunits, ribosomes, and polysomes can be isolated from detergent-treated cell extracts. In this study, we would perform polysome isolation deploying gene cassettes ENOD40p: HFGFP- RPL18 for primordial tissues and ENOD2p: HF-GFP-RPL18 for parenchymal tissues in Glycine max root nodules that express an epitope tagged version of ribosomal protein L18.
Over the last one decade, there has been several microarraysbased studies, which characterized transcriptional variations deployed in nodule formation. It has been embedded with couple of shortcomings, for instance; relative late time points study, incomplete representation of plant genes, discrimination of close paralogs, and reduced sensitivity. Lately, next generation sequencing technology have widened the horizon of transcription analyses in different legume species to detect symbiosis induced changes in late nodule developmental stages. Against this backdrop, we are accentuated to reveal early transcriptional changes induced in determinate type of soybean nodule by Bradyrhizobium japonicum.
In determinate type of nodule, two major nodule development zones are formed for instance, the Nodule Primordium (Npr) in the middle and it is encircled by nodule parenchyma (Npa). At later time point, the Npr converted to N-fixation zone and the Npa contained vascular bundles. Of these facts, it is not clear what early signaling pathways driving the conspicuous development of the nodule zones. In this context, mechanisms regulate the distinct gene expression profiles in Npr and Npa cell types has not understood clearly. The proposed research study is aimed at filling this knowledge gap by illustrating the molecular signatures that paves the way to cellular differentiation in root nodule development in soybean considering four different time points (5 dai, 7 dai, 10 dai & 14 dai).
The hypothesis is microRNAs (miRNAs) play important regulatory roles in spatio-temporal expression of their target genes during nodule developmental in soybean. For example, a gradient of microRNA localization between nodule primordium and parenchyma cells could result in distinct differentiation of these cell types. To test this hypothesis, one has to obtain both cell type-specific miRNA and transcriptome (miRNA target) profiles. Since, the majority of miRNA regulation occurs in the cytoplasm, we reasoned that comparison of nuclear and ribosomal transcriptome profiles would reveal genes whose expression is potentially regulated by post transcriptional mechanisms such as miRNA cleavage. Combining this information with cell type-specific miRNA profiles, and to test the above hypothesis and identify key miRNA-target pairs important for nodule cell differentiation. The use of Translating Ribosome Affinity Purification (TRAP) of nodule zone cells, namely from parenchyma and primordial tissues, to obtain cytoplasmic transcriptomes data.
Techniques to Determine Cell Type Specific Expression Profiles: TRAP Methods
TRAP is termed translating ribosome affinity purification, combines cell-type-specific transgene expression with affinity purification of translating ribosomes. It supersedes the need for tissue fixation, and facilitates to study the cell type-specific mRNA profiles of any genetically defined cell type. It has been successfully used to date in organisms ranging from D. melanogaster to mice, and human cultured cells. Multiple ribosomes or Polyribosomes (polysomes) are engaged in translation on a single mRNA. To evaluate the translation state of an mRNA, ribosomal subunits, ribosomes, and polysomes can be isolated from detergent-treated cell extracts.
In this study, the polysome isolation using gene cassettes ENOD40p: HF-GFP-RPL18 for primordial tissues, and ENOD2p: HF-GFP-RPL18 for parenchymal tissues in Glycine max root nodules that express an epitope tagged version of ribosomal protein L18 RPL18.
Relative abundance and differentially expressed mRNAs profile in two different tissue specific zones would help to understand the effect of regulatory role of miRNAs in cell differentiation and nodule development.
References
- Marie Turner et al. “Ectopic Expression of miR160 Results in Auxin Hypersensitivity, Cytokinin Hyposensitivity, and Inhibition of Symbiotic Nodule Development in Soybean”. Plant Physiology. 2013; 162: 2042-2055.
- Sponseller RA, et al. “Nitrogen dynamics in managed boreal forests: Recent advances and future research directions”. 2016; 45: 175-187.
- Johnson GV and DS Owens. “Fertilizer Nutrient Leaching and Nutrient Mobility: A Simple Laboratory Exercise.” J. Nat. Resour. Life Sci. Educ. 1996; 25: 128-131.
- Cheng Q. “Perspectives in biological nitrogen fixation research.” J Integr Plant Biol. 2008; 50: 786-798.
- Udvardi and Day. “Metabolite transport across symbiotic membranes of legume nodules.” Annual Review of Plant Physiology and Plant Molecular Biology. 1997; 48: 493-523.
- Celine Charon CJ, Eva Kondorosi, Adam Kondorosi and Martin Crespi. “enod40 induces dedifferentiation and division of root cortical cells in legumes.” Proc. Natl Acad. Sci. USA. 1997; 94: 8901-8906.
- Oldroyd GE, Downie JA. “Coordinating nodule morphogenesis with rhizobial infection in legume. Annual Review of Plant Biology. 2008; 59: 519-546.
- Kouchi H and Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 1993; 238: 106-119.
- Axtell MJ. “Classification and comparison of small RNAs from plants.” Annu Rev Plant Biol. 2013; 64: 137-159.
- Chapman EJ and JC Carrington. “Specialization and evolution of endogenous small RNA pathways.” Nat Rev Genet. 2007; 8: 884-896.
- Cuperus JT, et al. “Evolution and functional diversification of MIRNA genes.” Plant Cell. 2011; 23: 431-442.
- Wu L, et al. “DNA methylation mediated by a microRNA pathway.” Mol Cell. 2010; 38: 465-475.
- Manavella PA, et al. “Plant secondary siRNA production determined by microRNA duplex structure.” Proc Natl Acad Sci USA. 2012; 109: 2461-2466.
- Song JJ, et al. “Crystal structure of Argonaute and its implications for RISC slicer activity.” Science. 2004; 305: 1434-1437.
- Singh RK, et al. “Molecular evolution and diversification of the Argonaute family of proteins in plants.” BMC Plant Biol. 2015; 15: 23.
- Zhu H, et al. “Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development.” Cell. 2011; 145: 242-256.
- Xiang Liu TL, Yongchao Dou, Bin Yu, and Chi Zhang. “Identification of RNA silencing components in soybean and sorghum.” BMC Bioinform. 2014; 15: 4.
- Buxdorf K, et al. “Identification and characterization of a novel miR159 target not related to MYB in tomato.” Planta. 2010; 232: 1009-1022.
- Subramanian S, et al. “Novel and nodulation-regulated microRNAs in soybean roots.” BMC Genomics. 2008; 9: 160.
- Turner M, et al. “Genome organization and characteristics of soybean microRNAs.” BMC Genomics. 2012; 13: 169.
- Li H, et al. “Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation.” Plant Physiol. 2010; 153: 1759-1770.
- Zhe Yan MSH, Siwaret Arikit, Oswaldo Valdes-Lopez, Jixian Zhai, Jun Wang, Marc Libault, et al. “Identification of microRNAs and their mRNA targets during soybean nodule development: functional analysis of the role of miR393j-3p in soybean nodulation.” New Phytologist. 2015; 207: 748-759.
- Combier JP, et al. “MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula.” Genes Dev. 2006; 20: 3084-3088.
- Boualem A, et al. “MicroRNA166 controls root and nodule development in Medicago truncatula.” Plant J. 2008; 54: 876-887.