
Research Article
Austin J Plant Bio. 2023; 9(1): 1037.
Unveiling the Potential of OsRuvB DNA Helicases in Enhancing Salinity Stress Tolerance in Chickpea
Deepak Kumar1; Nita Lakra1*; Parul Sharma2; Annu Luhach1; Abbu Zaid3,4
1Department of Molecular Biology, Biotechnology and Bioinformatics CCS Haryana Agricultural University, India
2Department of Botany and Plant Physiology, CCS Haryana Agricultural University, India
3Plant Physiology and Biochemistry Section, Department of Botany, Aligarh Muslim University, Aligarh, India
4Department of Botany, GGM Science College, Cluster University Jammu, India
*Corresponding author: Nita Lakra Department of Molecular Biology, Biotechnology and Bioinformatics CCS Haryana Agricultural University, India. Email: nitahaubotany2019@gmail.com
Received: August 24, 2023 Accepted: October 09, 2023 Published: October 16, 2023
Abstract
Chickpea (Cicer arietinum L.) is a self-pollinated true diploid (2n=2x=16) cool season leguminous crop that ranks second among food grain legumes after soybean. It grows under a wide range of climatic conditions and is highly sensitive to salt stress. In the present study, transgenic chickpea plants (var. HC-1) carrying OsRuvB gene were screened for salt stress. Putative transformants were screened at an early stage through PCR amplification using gene specific primers and a transformation frequency of 36.2% was observed. Physio-biochemical analysis of selected T2 transgenic plants subjected to 100 mM salt stress showed that transgenic plants were able to maintain higher chlorophyll content, relative water content, cell viability, proline content, Na+/K+ content, catalase and peroxidase activity compared to the wild type plants. Whereas electrolytic leakage and lipid peroxidation were relatively less as compared to the wild type plants under 100 mM stress. Among all transgenic lines, line 8 performed well with respect to all the parameters studied and can be taken further for the development of transgenic chickpea plants for salt stress tolerance.
Keywords: Chickpea; Transgenic; OsRuvB; Physio-biochemical; Salt stress
Introduction
Chickpea (Cicer arietinum L.) is a self-pollinated true diploid (2n=2x=16) leguminous crop with a 738 Mbp genome that ranks second among food grain legumes in the world after soybean. It is grown in a wide range of environments in over 50 countries in subtropical and temperate regions of the world [1].
Although chickpea is grown in over 50 countries, 90% of the area under chickpea cultivation is in the developing countries, with southern and South-East Asian countries accounting for >79% of the global production [2]. Globally chickpea harvested area has been expanded from 9.63 million ha in 1980 to 12.65 million ha in 2016. Global chickpea production has also increased from 4.85 million tons in 1980 to 12.09 million tons in 2016. During 2017-18, a total of 25.23 million tons pulses were produced from 29.99 million ha. Out of total pulses production, a total of 112.29 lakh tons chickpea was produced from 105.61 lakh ha area which accounted for 35. 21 per cent and 44.50 per cent of area and production of total pulses, respectively [3]. The global growth rate of pulse production over the last decade has been 2.61%. In India, Madhya Pradesh is the highest chickpea producing state with a share of 41% of the national production. The other major chickpea producing states are Rajasthan, Maharashtra, Andhra Pradesh, Telangana, Uttar Pradesh, and Karnataka, which cover 95% the area under chickpea cultivation (State wise share to total production and area of chickpea in India 2015–2016). Cultivars grown in India are either native (desi) types or Mediterranean (Kabuli) types. The growth trends of area and production of pulses in Haryana found declining from 1970-71 to 2016-17. In 2017-2018 the production of chickpea went down from 36.4 thousand tons from 32 thousand ha of area [4].
Chickpea seeds consist of 19.3% protein, 64.6% carbohydrate, and vitamins [5]. Although chickpea has a high yield potential (4000 kg/ha), actual yields are quite low due to biotic and abiotic stresses [6]. High salinity is one of the major abiotic stress factors that reduce plant growth ultimately hindering crop productivity [7]. At least 20% of all irrigated lands are salt affected, with some estimates being as high as 50% [8]. Salinity is a soil condition characterized by a high concentration of soluble salts. Soils are classified as saline when ions concentration is such that osmotic pressure produced by ions are equivalent to that generated by 40 mM NaCl (i.e., 0.2 MPa) or higher [9]. Abiotic stresses affect various morphological, physiological, and biochemical processes, though all plants in a timely and well-coordinated response such that tolerant genotypes which are well adapted adaptation and survive under stress [10]. Excess of soluble salts in the soil leads to osmotic stress, resulting inion imbalances and toxicity, resulting in retarted plant growth [11,12]. Under stress conditions, Reactive Oxygen Species (ROS) are commonly generated and stored which cause oxidative damage to biomolecules such as lipids and proteins, resulting in cell death later in the process [13]. One of the approaches to overcome the consequence of salt stress is increasing salt tolerance in crop plants. Salinity is a complex trait which is associated with various cellular mechanisms [14].
Plant breeders have achieved some success in producing salt-tolerant lines/cultivars of some crops through conventional breeding; however, the main issue that conventional plant breeders have faced is the low magnitude of genetically based variation in the gene pools of most crop species. However, because of reproductive barriers, transferring salt-tolerant genes from wild relatives to domesticated crops is not easy [15] and thus very few examples of this approach being used effectively could be found in the literature [16,17]. The overall approach is time-consuming and labor-intensive; undesirable genes are frequently transferred alongside desirable ones; and reproductive barriers limit transfer of favorable alleles from inter-specific and inter-generic sources. Because of these factors, genetic engineering has emerged as an alternative strategy to conventional breeding for crop quality and impend yield potential in most crops. Nonetheless, plant biologists have focused heavily on genes that encode ion transport proteins, compatible organic solutes, antioxidants, heat-shock, and late embryogenesis abundant proteins as well as transcription factors for gene regulation to improve salt tolerance traits in various crops through genetic engineering [15]. Plants respond to stress by changing gene/transcript/protein expression levels at the molecular level. Overexpression of several stress-induced genes, including helicases, provides salinity stress tolerance in crop plants [18]. DNA helicases act as molecular proteins in a variety of cellular mechanisms and are required for nearly all DNA metabolic activities, including pre-mRNA splicing [19]. RuvB DNA helicases are capable of imparting salinity tolerance in Arabidopsis, rice, and pigeon pea [20]. RuvB is a member of AAA+ (ATPases Associated with diverse cellular Activities) superfamily, and part of SF6 superfamily which belongs to helicase class. Most of the helicases belong to DEAD-box protein superfamily. They are involved in regulation of cellular machinery such as DNA repair recombination, replication, transcription, translation initiation, ribosome biogenesis. So, they play a crucial role in stabilization of growth during stress conditions in plants. There are reports that helicases are up-regulated in response to abiotic stress in plants and help in survival under stressed conditions. Some of the examples of helicases which are activated under abiotic stresses are PDH45, PDH47, STRS1, STRS2, MCM6, p68 etc. [21-24]. The role of helicases has been established in various cellular functions such as replication, transcription, translation, gene regulation, DNA damage repair, chromatin remodeling and stress tolerance [25-28]. RuvB is a SF-6 type DNA helicase associated with diverse cellular activities such as protein folding, proteolysis, cytoskeleton regulation, and transcriptional control [29-32]. RuvB is well characterized in Escherichia coli [33], but now there are reports on characterization of RuvB in rice [34].
In the present study, Haryana chana 1 (HC-1) variety has been used to for the over expression of OsRuvB. HC-1 is a highly cultivated variety of chickpea used for commercial cultivation. India is the largest producer, consumer, and importer of pulse crops [35]. Due to the consumers and Government price support policies which are predominantly in favor of cereal crops it causes global production still lags behind [36]. Through given RuvB orthologous function in stress response, role in cellular functional reprogramming biotic stress we investigated its role in abiotic stress tolerance. To our knowledge, we were able to generate T2 chickpea heterologous expressing OsRuvB.
Materials and Methods
Materials
Plant Materials: In the present study chickpea variety, HC-1 and OsRuvB transgenic lines were used for studying morpho-physiological and biochemical responses of OsRuvB against salt stress. Seeds of chickpea were procured from Molecular Biology Biotechnology and Bioinformatics department, CCS HAU, Hisar.
OsRuvB, Plasmid and Agrobacterium tumefaciens strain: Agrobacterium tumefaciens strain: LBA4404 containing pCAMBIA1301 harboring OsRuvB gene was previously used for genetic transformation of HC-1 chickpea variety (Patent no. 252590) (Figure 1) [37]. The strain was a gift from Tuteja, N.K. ICGEB, Delhi.

Figure 1: pCAMBIA vector harboring OsRuvB.
Gene-specific primers: Gene-specific primers for OsRuvB gene used were designed using IDT software for PCR analysis. Both 557bp and 957bp primers were used for confirmation of OsRuvB (Table 1 & 2). These primers were synthesized from Eurofins Genomics, India Pvt. Ltd.
Primer
Sequence (5'→3')
Forward
GTGGCAGTGGGTGATGTTAT
Reverse
ATCTCAGTGGGACGGTATGT
Table 1: Gene specific primer (957 bp).
Primer
Sequence (5'→3')
Forward
CATCTCTCAGGAGCTAGGTAGT
Reverse
GATGTCTGTTGTCCGATCTCTC
Table 2: Gene specific primer (557 bp).
Genomic DNA Isolation
DNA from the young leaves of T2 and wild type chickpea pans were isolated using CTAB method [38]. DNA purification samples were re-extracted with equal volumes of phenol: chloroform: isoamylalcohol (25:24:1) followed by DNA precipitation and washing with 70% ethanol twice. DNA pellet was air dried, dissolved in appropriate volume of TE buffer and stored at -20oC till further use. The quality and quantity of isolated genomic DNA were estimated at 0.8% agarose gel electrophoresis.
RNase treatment: The RNA contamination in the samples was removed by adding 3 μl RNase A (10 mg/ml) to each sample and incubated at 37°C for 3-4 hours.
Purification of DNA: Samples were re-extracted with equal volumes of phenol:chloroform: isoamyl alcohol (25:24:1) followed by DNA precipitation and washing with 70% ethanol twice. DNA pellet was air dried, dissolved in appropriate volume of TE buffer and stored at -20°C till further use. The quality and quantity of isolated genomic DNA were estimated by 0.8% agarose gel electrophoresis.
Qualitative and quantitative estimation of genomic DNA: Quality of DNA was examined by submerged horizontal gel electrophoresis. A 0.8% (w/v) agarose gel was prepared for this (Sambrook et al., 1989). Gel casting plate was washed, air-dried and its ends were sealed with tape. Agarose was melted in 1 X TBE buffer and ethidium bromide was added at a concentration of 0.5 μg/ml of the gel solution. Gel solution was then poured into casting plate inserted with an appropriate comb to get 0.4-0.6 cm thick gel. Sealing tape was removed from both the ends as the gel solidified. The quality of DNA was examined by submerged horizontal gel electrophoresis. A 0.8% (w/v) agarose gel was used [39]. Samples were prepared by adding 4 μl of sterile distilled water, 2μl of 6X dye and 1μl of DNA sample (4:2:1). Samples were loaded in the wells and electrophoresis was carried out at a constant voltage (100 V) until dye migrated to the other end of the gel. Gel was then visualized under UV transilluminator, and photo was taken using UV Gel documentation system (Benchtop UVP). Quantitative estimation of genomic DNA was done by Nanodrop.
Molecular Analysis of Transgenic Chickpea Plants Carrying OsRuvB Gene
Screening of putative transgenic chickpea plants (HC-1) carrying OsRuvB gene: The putative transgenics were screened for the presence of OsRuvB gene through PCR using gene-specific primers. The gene-specific primer pair used in the present study was synthesized from Eurofins Genomics, India Pvt. Ltd.
PCR amplification: PCR reactions were carried out in 20μl reaction mixture containing 50ng DNA, 2μl of 10XPCR buffer (G-Biosciences) with MgCl2, 0.5μl of 10 mM of each forward and reverse primer (Eurofins), 0.5μl of 10mM dNTP (Thermo Scientific) and 2.5U Taq DNA polymerase (G-Biosciences). PCR was performed in Benchtop thermocycler.
Following PCR conditions were used: Initial denaturation 95°C for 10 min, denaturation 94°C for 1 min, annealing 52°C for 1.5 min, extension 72°C for 1.5 min and final extension 72°C for 10 min. Amplified products were stored at -20°C till further use.
Morpho-Physiological and Biochemical Analysis of Transgenic Plants under Salt Stress
Wild and transgenic plant morphology was recorded before and 7th Day after the abiotic stress treatment and photographed (Nikon D3500). Transgenic and non-transgenic chickpea plants were raised in dune sand pot and were subjected to 100 mM NaCl at flowering stage. Physiological and biochemical parameters relative water content, chlorophyll content, electrolyteleakage, lipid peroxidation, proline content, cell viability, sodium-potassium content, protein expression catalase and peroxidase activity were recorded at 7th day after salt treatment. WTNS - Wild Type under Non Stress, WTS - Wild Type under Stress, TNS - Transgenic under Non Stress, TS - Transgenic under Stress
Root morphology: The morphology of root was observed before and after stress and photograph taken.
Chlorophyll Content (mg/g FW): Leaf tissue was washed, blotted dry and dipped in test tubes containing 3 mL of DiMethyl Sulfoxide (DMSO) overnight as described by reference method [40]. Extracted chlorophyll in DMSO was estimated by recording its absorbance at 663 and 645 nm, respectively and its amount was calculated from the formula:
Chl. a = (12.3 A663–0.86 A645/ax1000xW) *V
Chl. b = (19.3 A645–3.6/ax1000xW) *V
Were, V: Volume of DMSO, A: Path length, W: Weight of Tissue
Relative Water Content (%): Leaf relative water content was calculated by using the method as described [41]. Leaf samples were collected and weighed immediately to measure weight. Leaves were placed separately in petri dishes filled with distilled water for 3 h. After, the same leaves (fully turgid) were weighed again and baked at 85oC for 72 h for drying and weighing and used to calculate percent Relative Water Content (RWC %).
RWC (%) = (Fresh Weight-Dry Weight/Turgid Weight-Dry weight)×100
Electrolyte leakage (%): Membrane injury was analyzed according to the standard method [42]. The electrolyte leakage was determined using a conductivity meter. One gram of fresh leaves was cut into pieces and placed in test tubes, in a water bath at 65oC for 1 hour and 20 ml of deionized water was added and kept overnight, the initial electrical conductivity of the medium, (i.e., EC1) was measured the following day.
Samples were then autoclaved for 20m for electrolyte release, cooled followed by m and final Electrical Conductivity, i.e. (EC2) was measured. Percent electrolyte leakage was calculated as follows:
Electrolyte leakage (%)=(1-EC1/EC2)×100
Lipid peroxidation (μmol/g FW): Lipid peroxidation was measured in terms of Malondialdehyde (MDA) content present in leaf tissues. MDA is a product of lipid peroxidation and was measured by Thiobarbituric Acid (TBA) [43]. Absorbance was read at 532 nm and 600 nm. The concentration of MDA was calculated using its extinction coefficient of 155 mM-1cm-1.
Cell viability assay: Cell viability assay was determined by Evans blue staining [44]. Evans blue release was estimated using a spectrophotometer (GENESYS 180 UV visible spectrometer) at 600 nm absorbance.
Proline content (μmol/g FW): Proline content was estimated by the reference method [45]. Proline content was calculated using a standard curve by taking absorbance at 520nm.
Catalase (μmole/g/min FW): Catalase (CAT) activity was estimated according to the reference method [46]. All steps of extraction were carried out at -4°C. Five hundred mg of leaves from transgenic and non-transgenic plants were used.
The leaves were washed with distilled water, filter paper dried and 3 mL extraction buffer (potassium phosphate) containing 0.1 mM EDTA, 1% (w/v) Polyvinylpyrrolidone (PVP, 0.5% Triton X-100 and 20% glycerol. pH was adjusted to 7.8. The homogenate was centrifuged at 10,000xg for 15 min at 4°C. The supernatant was aliquoted and used as crude enzyme extract. The reaction mixture in final volume of 3 mL, contained 0.1M phosphate buffer (pH 7.0), 10mM H2O2 and 50μL of cell-free extract. Reaction was initiated with the addition of H2O2. Enzyme activity was determined by via degradation of H2O2 at 240nm for 2 min. The enzyme activity was calculated using 39.4 mM-1cm-1 as the extinction coefficient value of H2O2. One unit (1U) of enzyme activity correlated with nmol H2O2 consumed during reading.
Peroxidase (μmole/g/min FW): Estimation of peroxidase (POX) activity was by reference method [47]. The extraction was the same as used for catalase. Three ml of reaction mixture contained 0.1 M phosphate buffer (pH 7.0), 0.1 mM guaiacol, 0.1 mM H2O2 and 50 μL cell-free extract. Reaction was started with H2O2. Absorbance at 470 nm was recorded for 2 min. The activity was calculated using the extinction coefficient value of 22.6 mM-1cm-1 for guaiacol. One unit of enzyme activity was equivalent to μmol of H2O2 oxidized.
Na+ - K+ (ppm) content: 500 mg plant material in a 50 or 100ml conical flask. Add 10 mL of diacid mixture of HNO3 and HClO4 in a ratio of 4:1 and kept overnight. Keep on a hot plate and heat gently first. Volume is reduced to 3-4 ml Cooled and transferred, diluted to 50mL using a volumetric flask and filtered for further analysis. After digestion, reading was read using a Flame photometer (S-935 Flame photometer) instrument.
Statistical analysis: Statistical analysis was carried out on physiological data recorded in T2 using two factorial CRD (Completely Randomized Design) test in OPSTAT program [48].
Results
Genomic DNA isolation
The agarose gel electrophoresis showed clear, sharp and intact bands with no shearing (Figure 2).

Figure 2: 0.8% agarose gel showing genomic DNA of T2 chickpea plants 1-33- T2 chickpea plant genomic DNA.
Molecular Characterization of Transgenic Chickpea Plants Carrying OsRuvB Hene
The putative transgenic plants were screened for the presence of OsRuvB gene in T2 generation with the help of PCR using gene-specific primers (Figure 3).

Figure 3: P: 1.5% agarose gel showing amplification of 557 bp fragment of OsRuvB gene in T2 generation plants. P: Positive Control (Plasmid DNA), NT: Non transformed, W: water, 7-12 represent T2 generation transgenic chickpea plants.
Screening of putative transgenic chickpea plants: An amplified fragment of 557 bp confirmed the presence of the transgene corresponding to the amplified product from plasmid DNA carrying OsRuvB gene. Out of 33 plants screened for the presence of OsRuvB gene, 12 plants showed a distinct band of 557 bp, representing a transformation efficiency of 36.6% (Table 3).
T1 Plants
Number of T2 Plant Screened
Confirmed by PCR
Line 4
15
5
Line 8
18
7
Table 3: Screening of T2 generation plants for the presence and integration of OsRuvB gene.
Morpho-Physiological Biochemical Parameters of Transgenic Plants Under Salinity
Morphology: A difference was seen in morphology of wild type and transgenic plants. Wild type plants had lesser growth in terms of shoots while transgenic plants show better growth. Morphology of plants before and 7th day after salt treatment is shown in Figure 4.

Figure 4: Morphology of plants before stress and 7th day after salt treatment.
Root Morphology: There was a clear difference observed in roots of wild and transgenic types under stress condition. Wild type under stress had poor root growth over wild type control. Transgenic under stress had more branched roots and looks more networks due to OsRuvBgene which carries more water absorbance over wild type (Figure 5).

Figure 5: Comparison of roots of wild and transgenic types under salinity. There was a clear difference observed in roots of wild and transgenic types under stress condition. Wild type under stress had poor root growth over wild type control.
Chlorophyll content (mg/g FW): The chlorophyll content of wild-type and transgenic plants were decreased under salt treatment (100mM). Minimum chlorophyll content was observed in wild type under stress. In wild type under stress there was a 0.7-fold decrease in chlorophyll content with respect to wild type control in normal conditions). We also observed a 1.23-fold increase in chlorophyll content in line 4 under stress with respect to wild type control. Line 4 transgenic control was also increased 1.29-foldwith respect to wild type control. In line 8, transgenic control there was a 1.6-fold increase of chlorophyll content with respect to wild type control whereas in line 8 transgenic under stress had increased1.4-fold respect to wild type Control. In both transgenic, line 8 was performing better under stress condition in terms of chlorophyll content (Figure 6, Table 4).

Figure 6: Effect of 100 mM salt stress on chlorophyll content of wild-type and T2transgenic chickpea plants.
Line
Chlorophyll Content (mg/g FW)
RWC (%)
EL (%)
Proline (μmole/g FW)
MDA (μmole/g FW)
Na+ (ppm)
K+ (ppm)
POX (μmole/g/min FW)
CAT (μmole/g/min FW)
WTNS
2.98±0.06
71.3±0.301
44.2±0.531
110±1.24
0.035±0.023
2.5±0.065
3.7±0.088
20.4±0.034
44.2±0.136
WTS
2.1±0.053
54.2±0.901
60.3±1.31
120±0.93
0.051±0.04
8.7±0.199
2.3±0.024
24.8±0.426
52.2±0.95
4LNS
3.86±0.024
79±1.692
40±0.77
150±0.156
0.028±0.07
2.1±0.04
4.7±0.051
23.56±0.76
57.2±0.774
4LS
3.68±0.073
70.3±0.256
50.2±0.82
180±1.592
0.044±0.002
6.1±0.079
2.1±0.014
33.21±0.553
60.62±0.537
8LNS
5.05±0.118
80.6±1.804
42.3±1.07
150±2.96
0.024±0.001
2.9±0.02
4.8±0.032
31.1±0.306
55.3±0.835
8LS
4.29±0.09
76.3±1.787
49.5±0.90
160±3.497
0.033±0.001
4.1±0.034
2.8±0.023
39.2±0.281
61.4±0.767
Table 4: Effect of 100mM NaCl salt treatment on various physio-biochemical parameters in T2 wild type and transgenic chickpea plants lines at 7th days after treatment.
Relative water content (%): Relative water content of wild type and transgenic lines were decreased under salt treatment (100mM). The minimum RWC was decrease in wild type under stress when compared with all other plants. There was a 0.7-fold decrease in RWC in wild type under stress condition. Inline 4 transgenic control there was an increase (1.1-fold) of RWC with respect to wild type control Line 4 under stress there was a decrease (0.9-fold) of RWC with respect to wild type control. In line 8 transgenic controls we observed a1.1-fold an increase of RWC and under stress it was 1.07-foldincrease compared to wild type control (Figure 7, Table 4).

Figure 7: Effect of 100mMsalt stress on relative water content of wild-type and T2transgenic chickpea plants.
Electrolyte leakage (%): Electrolyte leakage was more in wild type plant than the transgenic plant. There was a 1.3-fold increase of electrolyte leakage in wild type stress with respect to wild type control. In addition, line 4 control decreased (0.9-fold) whereas transgenic treated plants were increased by 1.1-foldcompared to wild type control. In line 8, transgenic control decreased 0.9-foldwhereas transgenic treated plants were increased 1.1-fold with respect to wild type control. Overall, line 8 had lesser electrolyte leakage than others (Figure 8, Table 4).

Figure 8: Effect of 100 mM salt stress on electrolyte leakage of wild-type and T2 transgenic chickpea plants.
Lipid peroxidation (μmol/g FW): Under stress conditions, lipid peroxidation increased in both transgenic and wild-type plants. The highest lipid peroxidation was observed in wild type under salt stress was increased 1.4-fold compared to wild type control. In line 4 transgenic control, it decreased 0.8-fold while in transgenic treated it was increased 1.2-fold over wild type control. In line 8 transgenic control lipid peroxidation decreases (0.6-fold) whereas in transgenic treated plants it was decrease (0.9-fold) fold. d (Figure 9, Table 4).

Figure 9: Effect of 100 mM salt stress on lipid peroxidation of wild-type and T2 transgenic chickpea plants.
Cell viability test: More cell viability was recorded in wild type control plants than wild type treated. Lower absorbance show less damage was observed in wild type, whereas wild type treated show more absorbance suggesting more cellular damage. Similar changes were observed in transgenic line (Figure 10).

Figure 10: Effect of salinity (100mM) on cell viability of wild and transgenic chickpea.
Proline content (μmol/g FW): Proline content of both transgenic and wild-type plants increased under stress conditions. The minimum proline content was observed in wild type control while under stress it was increased (1.09-fold). In line 4 transgenic controls were increased (1.3-fold) and treated also increased (1.6-fold) over wild type control. While in line 8 transgenic controls was increased (1.3-fold) and in transgenic treated it was increased (1.4-fold) over wild type control (Figure 11, Table 4).
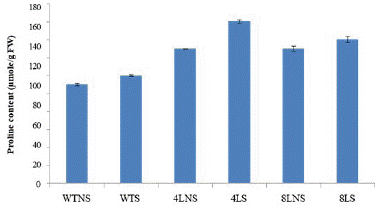
Figure 11: Effect of 100 mM salt stress on proline content of wild-type and T2 transgenic chickpea plants.
Catalase activity (μmole/g/min FW): Catalase (CAT) activity of both transgenic and wild-type plants increased under stress conditions. The minimum CAT activity was observed in wild type control. There was a1.1-fold increase in CAT activity in wild type under stress over wild type control. Among both transgenic lines, line 8 had more CAT activity. Line 4 transgenic control had a 1.2-fold increase in CAT activity whereas in transgenic treated it increased 1.37-fold over wild type control. Under stress line 8 had increased 1.3-fold while transgenic control was increased (1.2-fold) over wild type control (Figure 12, Table 4).

Figure 12: Effect of 100 mM salt stress on catalase activity of wild-type and T2 transgenic chickpea plants.
Peroxidase activity (μmole/g/min FW): Peroxidase activity in both wild type and transgenic plants increased under stress. The minimum peroxidase activity was observed in wild type control whereas in wild type under salt stress it increased 1.2-fold. Among both transgenic lines, line 4 control increased (1.1-fold) whereas those treated plants increased 1.6-fold compared to wild type control. Line 8 under stress had increased 1.9-fold whereas line 8 control had increased 1.5-fold over wild type control (Figure 13, Table 4).

Figure 13: Effect of 100 mM salt stress on peroxidase activity of wild-type and T2 transgenic chickpea plants.
Na+- K+ content: There was an increase of sodium content under salt stress in both wild type and transgenic type while potassium content was more in non-treated in both wild and transgenic type. There was a3.4-fold increase in sodium content in wild type under stress over wild type control. Along with this, line 4, transgenic control sodium levels decreased 0.8-fold, whereas transgenic treated increased 2.2-fold over wild type control. In line 8, transgenic control there was an increase 1.1-fold while in transgenic treated had a 1.6-fold increase over wild type control. While Na+/K+ ratio was higher under stress condition, K+/Na+ ratio is higher under non stress condition (Figure 14, Table 4).

Figure 14: Effect of 100mM salt stress on Na+ content of wild-type and T2 transgenic chickpea plants.
In the case of potassium ion, there was 0.6-fold decrease in wild type under stress over wild type control. In line 4 transgenic control, it was increased by 1.2-fold whereas in treated transgenics, had a 0.5-fold decrease over wild type control. In line 8 transgenic control, a 1.2-fold increase was observed, whereas in transgenic treated it was decreasing0.7-fold over wild type control. Overall, both transgenic lines had considerable Na+-K+ balance compared to their respective control (Figure 15-17, Table 4).

Figure 15: Effect of 100mM salt stress on K+ content of wild-type and T2 transgenic chickpea plants.

Figure 16: Ratio of Na+/K+ ion under stress of wild and transgenic chickpea plants.

Figure 17: Ratio of K+/Na+ ion under stress of wild and transgenic chickpea plants.
Protein study of wild type and transgenic plants: Protein content is increased under stress in both wild and transgenic plants. But it was increased in wild than transgenic. The minimum protein content was observed in wild type control. While the maximum protein content was observed in line 4 under stress. In wild type under stress protein content was increased (1.09-fold) over wild type. Along this in line 4 transgenic control the protein content was increased (1.2-fold) and in transgenic under stress it was increased (1.3-fold) over wild type control. In line 8 transgenic control had increased (1.1-fold) and in transgenic treated it was increased (1.2-fold) with respect to wild type control (Figure 18). The expression level of peptides of ~16, 20, 27, 37 and 66 kDa either showed decreased or increased accumulation with respect to WT. Less accumulation of Rubisco protein (~66kDa) under stress in both the WT and transgenic than their respective control was observed.

Figure 18: Effect of 100mM salt stress on protein content wild and transgenic types at 7th day after stress.
Discussion
In the present study, putative transformants had been examined for integration of OsRuvBin T2 generation with the use of PCR evaluation with gene-precise primers. Non-transformed plants served as negative control and plasmid DNA served as effective manipulate for screening of transgenics. Of 33 plants screened, 12 plants showed a 557 bp amplicon representing a transformation efficiency of 36.6%.
PCR study was undertaken with the genetic engineering salt-strain tolerance in V. mungo [49] via way of means of over-expression of the glyoxalase I (gly I) gene remoted from Brassica juncea [50]. Similarly, PCR primarily based screening of purported transgenic has been employed by varied researchers to substantiate the incorporation of the transgene in legumes like Cicer arietinum [51], Medicago sativa [52], Arabidopsis thaliana [53] and Glycine max [54].
Physiological biochemical evaluation of T2 generation plants subjected to 100mM NaCl at flowering stage to assess the transgene efficacy in mediating salt tolerance. Various physio-biochemical parameters such as chlorophyll content, relative water content, electrolyte leakage, lipid peroxidation, Na+-K+ content, root morphology, proline content, catalase and peroxidase, cell viability had been recorded.
A clear difference was seen in the morphology of wild and transgenic plants. Wild type plants show lesser growth in terms of growth parameters such as root and shoot while transgenic plants have better growth and development.
The chlorophyll content declined with salinity strain. The reduced chlorophyll content could be explained perhaps by the destabilizing effect of Na+ ions the thylakoid membrane by disrupting the lipid bilayer and membrane proteins. Chlorophyll content of the wild-type and transgenic plants were decreased under salt treatment (100 mM) than their respective control. NaCl treatment has been attributed to the destruction of chlorophyll pigments and the instability of pigment-protein complex [55-57].
The relative water content is a physiological index that is used to investigate the water retention capacity and serves as a suitable parameter to measure the water status and the osmotic settings of plants under abiotic stress [58]. RWC decreased in both WT and transgenic lines under salt stress, but the decrease was more pronounced in WT plants, suggesting that as the duration of salt stress increased, transgenic lines were effectively able to retain more water in their tissues than WT. The results were comparable to related transformation studies that reported better water retention in in transgenic plants such as corn [59], tomato [60], and tobacco [61] for drought stress [62] and pigeon pea [63] for salt stress.
The degree of cell membrane damage induced by salt stress can be measured by estimating the electrolyte leakage. Electrolyte leakage is considered a physiological indicator of tolerance to salt. In the present investigation the wild and transgenic both showed an increase of electrolyte under stress. But transgenic plants were less damaged over wild type. They were more tolerant under stress conditions than wild type. In a study, a defensin gene, Ca-AFP, from Cicer arietinum, was cloned and transformed in Arabidopsis thaliana and under simulated water-deficit conditions, the transgenic A. thaliana plants had higher accumulation of the Ca-AFP transcript compared to that under non-stress condition and exhibited reduced ion leakage in the transgenic plants as compared to wild-type plants [64].The transgenic peanut plants not only accumulated high levels of solutes, but also showed increased membrane integrity under severe stress conditions. Transgenic peanut lines over-expressing HDG11 showed significantly reduced electrolyte leakage [65]. Similar results were obtained in soybean over-expressing MsWRKY11 [66], tobacco over-expressing p68 [19]. NaCl toxicity, the foremost form of salt in most saline soils, enhances the Na content and therefore influences the absorption of different mineral factors [67]. Differences in the accumulation of K+/ Na+ can also be involved in cultivar behavior under salt stress [68] which suggests that toxic ions can accumulate more in genotypes due to increased perspiration. Sensitive varieties accumulate ions more quickly than tolerant and this accumulation of ions leads to the death of the leaves and gradually to the death of the plant [69].
In the present investigation the sodium content was high in stress condition and potassium vice-versa. The maximum sodium content was present in wild type whereas in transgenic lines it accumulated less over wild type and potassium content vice-versa. Overall, the transgenic line imparts better tolerance in ion toxicity than wild type. Similar results were found in pea at different salt treatments where leaf K+ /Na+ ratio decreased significantly with increasing salt levels [70]. It has been reported in the youngest fully developed leaves that highest Na+ concentration was related to a decrease in chickpea yield under salty conditions [71].
Additional evidence for decreased oxidative damage was tested through much less membrane harm index and MDA content inside the transgenic plants as compared to chickpea plants. MDA is an indicator of peroxide membrane lipid, so it is an effective marker of cell damage induced by oxidative stress. Reduce MDA levels to control oxidative stress and reduce the rate of oxidative stress membrane damage It can be concluded from this that the OsRuvB transgene plays a key role in maintaining a lower ROS content under salt stress conditions and thus prevents membrane damage in plants. In this study the MDA content was maximum in wild type under stress while transgenic had also increased MDA content but it was low as compared to wild type.
Same trends were observed in pigeon pea have been reported [72] and in rice [73]. Proline is associate degree osmoprotectant that incorporates a key role in maintaining the diffusion balance in crop plants, protective the cell organelles, enzymes and enhancing the osmolarity of the plant cells below stress conditions [72]. In present investigation proline content was increase in both wild and transgenic. Accumulation under salt stress was comparatively lower in wild-type plants as compared to transgenic plants. Among transgenic lower accumulation of proline were observed in line 8 then line 4. However, the increase of proline and total soluble sugars in wild-type plants was significantly higher than that of transgenic plants under stress which indicates that other osmotic compounds such as glycine, betaine or polyamines, etc., not considered in this study, can play a decisive role in osmotic protection [74].
Excessive Reactive Oxygen Species (ROS) formation can induce oxidative stress, leading to cell damage that can culminate in cell death. Therefore, cells have antioxidant (catalase, peroxidase etc.) networks to scavenge excessively produced ROS. Catalase is a heme-containing enzyme that catalyzes the breakdown of H2O2 into H2O and O2. Catalase eliminates the H2O2 generated in peroxisomes by oxidases involved in Β-oxidation of fatty acids, photorespiration, purine catabolism and during oxidative stress. A significantly higher catalase activity in transgenic lines of chickpea demonstrated their ability to scavenge ROS. In the present study, CAT was increased in all plants. But more pronounced CAT activity was seen in transgenic line. In transgenic line 4 had more CAT activity which shows greater tolerance under stress. Improved catalase activity had been cited in Arabidopsis [75] and peanut [76].
Peroxidase is a heme-containing protein, which oxidizes certain substrates at the expense of H2O2 and rid the cellular of excess peroxide produced with the aid of metabolic interest under both regular and pressure conditions. The production of antioxidant enzymes under salt stress enables an efficient management of ROS and therefore improves the life of the cells and their components. In the present study, peroxidase activity was increased in both wild and transgenic types under 100 mM salt stress. POX activity was recorded low in wild while high in transgenic. Among transgenic line 8 had more POX activity. A similar trend was observed in Sorghum bicolor [77,78] and tomato [79].
Plant protein pattern is influenced by salinity in two ways. First one it lowers total protein content [80] and second is restricting the production of specific proteins [81] required for tolerating effects of salinity through engaging ABA [82]. Total protein content was higher in both wild and transgenic under stress condition but more in transgenic case.
Confirmed with our findings, enhanced protein content upon salt stress is reported in different tolerant plant species [83,84]. Stress-regulated proteins could be classified into two groups: proteins that take part in signal transduction comprising transcription factors, RNA-binding proteins, protein kinases and phosphatases [85,86] and proteins that might be directly playing role in plant survival under stress conditions. The second group includes proteins involved in ion homoeostasis through increased synthesis of osmolytes and compatible solutes [87] and these are rectified through induction in water channels [88], oxygenic enzymes system [89] and may be some specific protective proteins [90]. Therefore, understanding the molecular details of plant stress response depends on clarifying the biological activity of individual proteins and their interaction with other cellular components. Salt stress proteins of low mass namely 20-24 and 26 kDa have been reported in barley [91,92]. The increase level of 26 and 27 kDa proteins in barley by salt stress [93] and in rice considered to be the acquisition of tolerance to salt stress since the amount of 26 kDa protein concentration of NaCl in the medium [94].
Conclusions
Chickpea is a self-pollinated true diploid (2n=2x=16) winter season leguminous crop that ranks second among food grain legumes in the world after soybean. Presently, it is cultivated in more than fifty countries across the Indian subcontinent, North Africa, the Middle East, southern Europe, USA, and Australia. India is the largest producer of chickpea accounting for 75% of the global chickpea production. Conventional breeding has made numerous attempts to enhance chickpea towards salt stress however no predominant step forward has been taken. Introduction of foreign genes for salt tolerance from primitive landraces and elite cultivars is a dependable opportunity and may be achieved with the assist of genetic engineering.
In the present study, transgenic chickpea plants (var. HC-1) carrying OsRuvB gene were screened for salt stress tolerance. The seeds of wild and chickpea were grown in pots in green house. After germination leaves were collected for DNA isolation and run gel electrophoresis for confirmation of DNA. PCR based screening was used for identification of putative transformed plants using OsRuvB gene-specific primers which produced an amplicon size of 557 bp. Out of 33 plants screened for the presence of OsRuvB gene, 12 plants showed a distinct band of 557 bp, representing a transformation efficiency of 36.6%.
Transgenic and wild type T2 chickpea plants were subjected to salt stress (100 mM) after germination at flowering stage and various physio-biochemical parameters like relative water content, chlorophyll content, electrolyte leakage, Na+-K+ content, lipid peroxidation, root morphology, catalase activity, peroxidase activity and proline content were studied.
Salt stress affected the various physio-biochemical parameters resulting in decrease in chlorophyll and relative water content and an increase in electrolyte leakage, lipid peroxidation and proline content. The activity of antioxidant enzymes, catalase and peroxidase increased with salt stress. Sodium content is increased and potassium vice versa.
Among all the transgenic lines, line 8 performed better in terms of various physio- biochemical parameters except ROS in which line 4 performed better studied under salt stress conditions.
From the present study we can conclude that OsRuvB coding for DNA helicase in mitigating salt stress in transgenic chickpea plants. Field studies are required to further confirm the effectiveness of the chickpea transgenic plants in the real saline situations.
Author Statements
Author Contributions
Conceptualization, N.L. and D.K.; methodology, D.K.; validation, N.L., and A.Z.; formal analysis, D.K; P.S.; investigation, D.K; and. N.L.; resources, A.Z.; data curation, D.K.; and P.S. writing—original draft preparation, D.K; N.L; P.S; and A.Z; writing—review and editing, A.Z; N.L; and P.S.; visualization, D.K.; supervision, N.L; project administration, N.L. All authors have read and agreed to the published version of the manuscript.
References
- Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, et al. Draft genome sequence of chickpea (Cicer arietinum L.) provides a resource for trait improvement. Nat Biotechnol. 2013; 31: 240-6.
- Gaur PM, Thudi M, Samineni S, Varshney. Advances in chickpea genomics. Legumes Omic Era. 2014: 73-94.
- Qasim A, Kumar V, Mehta VP. Structural dynamics of Agri-import-export of pulse crops to the total agriculture trade in India. Legume Res Int J. 2021; 44: 1449-54.
- Nimbrayan PK, Bhatia JK. Growth and Instability in Area, Production and Productivity of pulses in Haryana vis-à-vis India. Growth. 2019; 35: 1-8.
- Abu-Salem FM, Abou-Arab EA. Physico-chemical properties of tempeh produced from chickpea seeds. J Am Sci. 2011; 7: 107-18.
- Canci H, Toker C. Evaluation of yield criteria for drought and heat resistance in chickpea (Cicer arietinum L.). J Agron Crop Sci. 2009; 195: 47-54.
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012; 1819: 97-103.
- Pitman MG, Lauchli A. Global impact of salinity and agricultural ecosystems. Salinity Environ-Plants-Mol. 2002: 3-20.
- Brown Jr GE. Research databases. Bibliography on salt tolerance. ARS: United States Department of Agriculture. Riverside: U S Department of Agriculture Research and Services. 2008; 8908.
- Lakra N, Kaur C, Singla-Pareek SL, Pareek A. Mapping the early salinity response triggered proteome adaptation in contrasting rice genotypes using iTRAQ approach. Rice. 2019; 12: 1-22.
- Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002; 25: 239-50.
- Rout NP, Shaw BP. Salt tolerance in aquatic macrophytes: ionic relation and interaction. Biol Plant. 2001; 44: 95-9.
- Molassiotis A, Sotiropoulos T, Tanou G, Diamantidis G, Therios I. Boron-induced oxidative damage and antioxidant and nucleolytic responses in shoot tips culture of the apple rootstock EM 9 (Malus domestica Borkh). Environ Exp Bot. 2006; 56: 54-62.
- Shad MA, Nawaz H, Rehman T, Ikram N. Determination of some biochemicals, phytochemicals and antioxidant properties of different parts of Cichorium intybus L.: A comparative study. J Anim Plant Sci. 2013; 23: 1060-6.
- Ashraf M, Athar HR, Harris PJC, Kwon TR. Some prospective strategies for improving crop salt tolerance. Adv Agron. 2008; 97: 45-110.
- Ashraf MY, Wu L. Breeding for salinity tolerance in plants. Crit Rev Plant Sci. 1994; 13: 17-42.
- Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004; 55: 307-19.
- Amin M, Elias SM, Hossain A, Ferdousi A, Rahman MS, Tuteja N et al. Over- expression of a DEAD-box helicase, PDH45, confers both seedling and reproductive stage salinity tolerance to rice (Oryza sativa L.). Mol Breeding. 2012; 30: 345-54.
- Tuteja N, Banu MSA, Huda KMK, Gill SS, Jain P, Pham XH et al. Pea p68, a DEAD-box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLOS ONE. 2014; 9: e98287.
- Holt III BF, Boyes DC, Ellerström M, Siefers N, Wiig A, Kauffman S et al. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev Cell. 2002; 2: 807-17.
- Pham XH, Reddy MK, Ehtesham NZ, Matta B, Tuteja N. A DNA helicase from Pisum sativum is homologous to translation initiation factor and stimulates topoisomerase I activity. Plant J. 2000; 24: 219-29.
- Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci USA. 2005; 102: 509-14.
- Vashisht AA, Pradhan A, Tuteja R, Tuteja N. Cold-and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 2005; 44: 76-87.
- Kant P, Kant S, Gordon M, Shaked R, Barak S. STRESS RESPONSE SUPPRESSOR 1 and STRESS RESPONSE SUPPRESSOR 2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 2007; 145: 814-30.
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989; 17: 4713-30.
- Nath M, Garg B, Sahoo RK, Tuteja N. PDH45 overexpressing transgenic tobacco and rice plants provide salinity stress tolerance via less sodium accumulation. Plant Signal Behav. 2015; 10: e992289.
- Tuteja N, Ahmad P, Panda BB, Tuteja R. Genotoxic stress in plants: shedding light on DNA damage, repair and DNA repair helicases. Mutation research/ reviews in mutation research. 2009; 681: 134-49.
- Tuteja N, Tarique M, Trivedi DK, Sahoo RK, Tuteja R. Stress-induced Oryza sativa BAT1 dual helicase exhibits unique bipolar translocation. Protoplasma. 2015; 252: 1563-74.
- Sakato M, King SM. Design and regulation of the AAA+ microtubule motor dynein. J Struct Biol. 2004; 146: 58-71.
- Rotanova TV, Botos I, Melnikov EE, Rasulova F, Gustchina A, Maurizi MR, et al. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 2006; 15: 1815-28.
- Schumacher J, Joly N, Rappas M, Zhang X, Buck M. Structures and organization of AAA+ enhancer binding proteins in transcriptional activation. J Struct Biol. 2006; 156: 190-9.
- Mueller-Cajar O. The diverse AAA+ machines that repair inhibited RuBisCO active sites. Front Mol Biosci. 2017; 4: 31.
- Tsaneva IR, Illing G, Lloyd RG, West SC. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992; 235: 1-10.
- Saifi SK, Passricha N, Tuteja R, Nath M, Gill R, Gill SS, et al. OsRuvBL1a DNA helicase boost salinity and drought tolerance in transgenic indica rice raised by in planta transformation. Plant Sci. 2023; 335: 111786.
- FAOSTAT, Food and Agriculture Organization of the United Nations statistics. Available from: http://www.fao.org/faostat/.
- Merga B, Haji J. Economic importance of chickpea: production, value, and world trade. Cogent Food Agric. 2019; 5: 1615718.
- Khatodia S, Kharb P, Batra P, Chowdhury VK. Development and characterization of transgenic chickpea (Cicer arietinum L.) plants with cry1Ac gene using tissue culture independent protocol. Int J Adv Res. 2014; 2: 323-31.
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RWL. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984; 81: 8014-8.
- Sambrook J, Fritsch ER, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 1989.
- Sawhney V, Singh DP. Effect of chemical desiccation at the post-anthesis stage on some physiological and biochemical changes in the flag leaf of contrasting wheat genotypes. Field Crops Res. 2002; 77: 1-6.
- Smart RE, Bingham GE. Rapid estimates of relative water content. Plant Physiol. 1974; 53: 258-60.
- Dionisio-Sese ML, Tobita S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998; 135: 1-9.
- Heath RL, Packer LJ. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968; 125: 189-98.
- Sanevas N, Sunohara Y, Matsumoto H. Characterization of reactive oxygen species-involved oxidative damage in Hapalosiphon species crude extract-treated wheat and onion roots. Weed Biol Manage. 2007; 7: 172-7.
- Bates P, Bradley WE, Glen E. First report on the standardization of terminology of lower urinary tract [unction. Urinary incontinence. Procedures related to the evaluation of urine storage: cystometry, urethral closure pressure profile, units of measurement. Br J Urol. 1976; 48: 3942.
- Aebi H. Catalase in vitro. Methods Enzymol. 1984; 105: 121-6.
- Siegel SM, Siegel BZ. Peroxidase activity and stress: a complex relationship. Mol Physiol Aspects Plant Peroxidases. 1986: 427-31.
- Sheoran OP, Tonk DS, Kaushik LS, Hasija RC, Pannu RS. Statistical software package for agricultural research workers. Recent advances in information theory, statistics and computer applications. DS Hooda and RC Hasija department of mathematics statistics. Hisar: CCS HAU. 1998; 139-43.
- Bhomkar P, Upadhyay CP, Saxena M, Muthusamy A, Shiva Prakash N, Pooggin M, et al. Salt stress alleviation in transgenic Vigna mungo L. Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum yellow leaf curling virus (CmYLCV) promoter. Mol Breeding. 2008; 22: 169-81.
- Veena VS, Reddy VS, Sopory SK. Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 1999; 17: 385-95.
- Kiran KGS, Sujata KG, Kumar BMV, Karba NN, Reddy KJ, Rao MS, et al. Heterologous expression of P5CS gene in chickpea enhances salt tolerance without affecting yield. Biol Plant. 2011; 55: 634-40.
- Tang L, Cai H, Zhai H, Luo X, Wang Z, Cui L, et al. Overexpression of Glycine soja WRKY20 enhances both drought and salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Cell Tissue Organ Cult (PCTOC). 2014; 118: 77-86.
- Sun X, Luo X, Sun M, Chen C, Ding X, Wang X, et al. A glycine soja14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014; 55: 99-118.
- Ning W, Zhai H, Yu J, Liang S, Yang X, Xing X, et al. Overexpression of Glycine soja WRKY20 enhances drought tolerance and improves plant yields under drought stress in transgenic soybean. Mol Breeding. 2017; 37: 19.
- Jaleel CA, Sankar B, Sridharan R, Panneerselvam R. Soil salinity alters growth, chlorophyll content, and secondary metabolite accumulation in Catharanthus roseus. Turk J Biol. 2008; 32: 79-83.
- Taïbi K, Taïbi F, Ait Abderrahim LA, Ennajah A, Belkhodja M, Mulet JM. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defense systems in Phaseolus vulgaris L. S Afr J Bot. 2016; 105: 306-12.
- Sayyad-Amin P, Jahansooz MR, Borzouei A, Ajili F. Changes in photosynthetic pigments and chlorophyll-a fluorescence attributes of sweet-forage and grain sorghum cultivars under salt stress. J Biol Phys. 2016; 42: 601-20.
- Ravikumar G, Manimaran P, Voleti SR, Subrahmanyam D, Sundaram RM, Bansal KC, et al. Stress-inducible expression of AtDREB1Atranscription factor greatly improves drought stress tolerance in transgenic indica rice. Transgen Res. 2014; 23: 421-39.
- Lu Y, Li Y, Zhang J, Xiao Y, Yue Y, Duan L, et al. Overexpression of Arabidopsis molybdenum cofactor sulfurase gene confers drought tolerance in maize (Zea mays L.). PLOS ONE. 2013; 8: e52126.
- Rai AC, Singh M, Shah K. Engineering drought tolerant tomato plants over-expressing BcZAT12 gene encoding a C2H2 zinc finger transcription factor. Phytochemistry. 2013; 85: 44-50.
- Liu X, Hua X, Guo J, Qi D, Wang L, Liu Z, et al. Enhanced tolerance to drought stress in transgenic tobacco plants overexpressing VTE1 for increased tocopherol production from Arabidopsis thaliana. Biotechnol Lett. 2018; 30: 1275-80.
- Yadav NS, Shukla PS, Jha A, Agarwal PK, Jha B. The. SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2018; 12: 1-18.
- Surekha CH, Kumari KN, Aruna LV, Suneetha G, Arundhati A, Kavi Kishor PB. Expression of the Vigna aconitifolia P5CSF129A gene in transgenic pigeon pea enhances proline accumulation and salt tolerance. Plant Cell Tiss Organ Cult. 2014; 116: 27-36.
- Kumar M, Yusuf MA, Yadav P, Narayan S. Overexpression of chickpea defensin gene confers tolerance to water-deficit stress in Arabidopsis thaliana. Front Plant Sci. 2018; 10: 290.
- Banavath JN, Chakradhar T, Pandit V, Konduru S, Guduru KK, Akila CS, et al. Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea L.). Front Chem. 2018; 6: 34.
- Wang Y, Jiang L, Chen J, Tao L, An Y, Cai H, et al. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLOS ONE. 2018; 13: e0192382.
- Greenway H, Munns R. Mechanisms of salt tolerance in non-halophytes. Annu Rev Plant Physiol. 1980; 31: 149-90.
- Zheng Y, Wang Z, Sun X, Jia A, Jiang G, Li Z. Higher salinity tolerance cultivars of winter wheat relieved senescence at reproductive stage. Environ Exp Bot. 2008; 62: 129-38.
- Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002; 25: 239-50.
- Sanwal SK, Kumar A, Mann A, Kaur G. Differential response of pea (Pisum sativum) genotypes exposed to salinity in relation to physiological and biochemical attributes. Indian J Agri Sci. 2018; 88: 149-56.
- Turner NC, Colmer TD, Quealy J, Pushpavalli R, Krishnamurthy L, Kaur J, et al. Salinity tolerance and ion accumulation in chickpea (Cicer arietinum L.) subjected to salt stress. Plant Soil. 2013; 365: 347-61.
- Surekha CH, Kumari KN, Aruna LV, Suneetha G, Arundhati A, Kavi Kishor PB. Expression of the Vigna aconitifolia P5CSF129A gene in transgenic pigeon pea enhances proline accumulation and salt tolerance. Plant Cell Tiss Organ Cult. 2014; 116: 27-36.
- Sahoo RK, Ansari MW, Tuteja R, Tuteja N. OsSUV3 transgenic rice maintains higher endogenous levels of plant hormones that mitigates adverse effects of salinity and sustains crop productivity. Rice (N Y). 2014; 7: 17.
- Ahmad P, Jaleel CA, Sharma S. Antioxidant defense system, lipid peroxidation, proline- metabolizing enzymes, and biochemical activities in two Morus alba genotypes subjected to NaCl stress. Russ J Plant Physiol. 2010; 57: 509-17.
- Zhao X, Wei P, Liu Z, Yu BX, Shi H, Na S, et al. Antiporter GmsSOS1 enhances antioxidant enzyme activity and reduces Na+ accumulation in Arabidopsis and yeast cells under salt stress. Acta Physiol Plant. 2017; 39: 1-11.
- Singh N, Mishra A, Jha B. Over-expression of the peroxisomal ascorbate peroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar Biotechnol (NY). 2014; 16: 321-32.
- Anjaneyulu E, Reddy PS, Sunita MS, Kishor PBK, Meriga B. Salt tolerance and activity of antioxidative enzymes of transgenic finger millet overexpressing a vacuolar H+- pyrophosphatase gene (SbVPPase) from Sorghum bicolor. J Plant Physiol. 2014; 171: 789-98.
- Surender Reddy P, Jogeswar G, Rasineni GK, Maheswari M, Reddy AR, Varshney RK, et al. Prolineover-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol Biochem. 2015; 94: 104-13.
- Rai AC, Singh M, Shah K. Engineering drought tolerant tomato plants over-expressing BcZAT12 gene encoding a C2H2 zinc finger transcription factor. Phytochemistry. 2013; 85: 44-50.
- Delgado MJ, Garrido JM, Ligero F, Lluch C. Nitrogen fixation and carbon metabolism by nodules and bacteroids of pea plants under sodium chloride stress. Physiol Plant. 1993; 89: 824-9.
- Chen CCS, Plant AL. Salt-induced protein synthesis in tomato roots: the role of ABA. J Exp Bot. 1999; 50: 677-87.
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988; 39: 439-73.
- Amini F, Ehsanpour AA. Soluble proteins, proline, carbohydrates and Na+/K+ changes in two tomato (Lycopersicon esculentum Mill.) cultivars under in vitro salt stress. Am J Biochem Biotechnol. 2005; 1: 212-6.
- Najaphy A, Khamssi NN, Mostafaie A, Mirzaee H. Effect of progressive water deficit stress on proline accumulation and protein profiles of leaves in chickpea. Afr J Biotechnol. 2010; 9: 7033-6.
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000; 3: 217-23.
- Xiong L, Zhu JK. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2002; 25: 131-9.
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000; 12: 431-4.
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. The role of aquaporins in cellular and whole plant water balance. [Biochimica et biophysica acta (BBA)]. Biochim Biophys Acta. 2000; 1465: 324-42.
- Bohnert HJ, Sheveleva E. Plant stress adaptations—making metabolism move. Curr Opin Plant Biol. 1998; 1: 267-74.
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol. 2002; 51: 463-99.
- Ramagopal S. Salinity stress induced tissue-specific proteins in barley seedlings. Plant Physiol. 1987; 84: 324-31.
- Hurkman WJ, Tao HP, Tanaka CK. Germin-like polypeptides increase in barley roots during salt stress. Plant Physiol. 1991; 97: 366-74.
- Hurkman WJ, Tanaka CK. The effects of salt on the pattern of protein synthesis in barley roots. Plant Physiol. 1987; 83: 517-24.
- Shirata K, Takagishi H. Salt-induced accumulation of 26 and 27-kD proteins in cultured cells of rice plant. Soil Sci Plant Nutr. 1990; 36: 153-7.