
Research Article
Austin J Proteomics Bioinform & Genomics. 2022; 7(1): 1033.
Pharmacokinetics and Molecular Docking Studies of Sinapaldehyde and MCM7 Protein against Meier-Gorlin Syndrome
Barman AD* and Ghosh A
School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, India
*Corresponding author: Archita Dev Barman, School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, India
Received: March 17, 2022; Accepted: April 11, 2022; Published: April 18, 2022
Abstract
Objective: The objective of this study to investigate the MCM7 protein in contrast to Meier-Gorlin Syndrome which contains most of the ORC1, ORC4, ORC6, CDT1, and CDC6 causative genes in the preinitiation complex disruptions.
Methods: Sinapaldehyde and MCM7 were retrieved from Protein Data bank and PubChem database in the format of PDB and SDF format. Further, SDF format was converted to PDB format using Biovia Discovery Studio visualizer. Sinapaldehyde was subjected to pharmacoinformatics study as well as Molecular Docking analysis to find out the best anti-Gorlin potential.
Results: Sinapaldehyde showed the -5.9kcal/mol binding affinity against MCM7 protein which considers to be a better candidate drug molecule against Meier-Gorlin Syndrome and validated using Pharmacokinetics mechanism.
Conclusion: Molecular Docking studies and ADMET analysis showed the inhibitory activity of Sinapaldehyde with respect to Meier-Gorlin Syndrome and related pathway analysis and can be better compound for in vitro studies for future perspectives.
Keywords: MCM7; ADMET; Sinapaldehyde; Meier-Gorlin Syndrome
Introduction
Meier-Gorlin Syndrome (MGS) is an autosomal recessive syndrome characterized by short stature, growth retardation, small mouth, small ears and ear canals, microcephaly, patellar anomalies like small kneecaps [1]. Meier-Gorlin syndrome is a kind of primordial dwarfism and it is strongly associated with the preinitiation complex disruption. Few other symptoms include small head size, problem in feeding, abnormalities in the respiratory tract. Females who have this syndrome may have underdeveloped breasts. The mutated genes (ORC1, ORC4, ORC6, CDT1, and CDC6) encoding constituents of the pre-replication complex (PRC) are found to be one of the primary causes in patients with MGS [1]. In most cases, this syndrome is inherited to the child as an autosomal recessive pattern. Diagnosis of some major diseases can be challenging, even the doctors for such cases look at the patient’s medical history, what symptoms the patient has, the laboratory results, etc. to diagnose that patient. Based on the symptoms and signs, MGS is diagnosed through genetic testing. Most people, who are affected by MGS, can expect normal life longevity. In rare cases, the severity of this syndrome is too harsh such as abnormal breathing in infancy or later in life and the life span of patient also depends on how severe the disease is [2]. MCM7 helicase is a heterohexameric complex which has some essential roles and in the early stages of DNA replication processes it is a part of both prereplication and pre-initiation complexes. It is confirmed that variants in MCM7 protein are deleterious and through interfering with the MCM complex formation, it has an impact inefficiency of S phase progression. The MCM7 protein is encoded by the MCM7 gene which is one of the minichromosome maintenance proteins which is highly responsible for the eukaryotic genome replication. Replication fork is formed by the hexameric protein complex. The CDK4 associated with this protein regulates the binding of MCM7 with the tumour suppressor protein RB1 [3].
Materials and Methods
Preparation of ligand
The required information and structures of our ligand, sinapaldehyde, were retrieved from the PubChem database which was later used for molecular docking simulations with the receptor MCM7 [9-11]. The structures were downloaded from PubChem in Standard Data Format (SDF) which was converted into Protein Data Bank (PDB) format using PyMol version 2.5.1 [4-6].
Receptors
We have used MCM7 receptors against the selected ligand sinapaldehyde to inhibit the activity of Mier-Gorlin Syndrome.
Homology modelling
We have used SWISS-MODEL for the homology modelling of the MCM7 receptor [12-14]. The FASTA sequence of the receptor was retrieved from the NCBI database to prepare the template model [15,16]. A total of 8 models were retrieved from the server. We have selected the template (Model 01(MCM7)) which had a 100% sequence identity with respect to the chosen template.
ADMET analysis
We have used the drug discovery tool, SwissADME for the analysis of ADMET, physicochemical properties, pharmacokinetics and drug likeliness. The SwissADME analysis is based on the “Five Rules of Lipinski” which follows some standard criteria such as molecular mass, lipophilicity, hydrogen bond acceptors and donors. This was performed to check the oral feasibility of the drug in humans [17,18].
Boiled-egg
SwissADME server was used for the Boiled-Egg structure analysis. The Boiled-Egg plot is used to evaluate the gastrointestinal absorption and brain penetration through the Blood-Brain Barrier of the drug. The white zone indicates a high likelihood of passive absorption through the gastrointestinal system, while the yellow region (yolk) indicates a high likelihood of brain penetration [17-19].
Molecular docking analysis
Molecular docking was performed using virtual software PyRx [7,8]. The purpose behind performing molecular docking is to predict the possible interactions of the sinapaldehyde with the targeted MCM7 protein receptor. PyRx, which is virtual screening software, uses Vina and AutoDock 4.2 as docking software from which the binding affinity and the Root Mean Square Deviation (RMSD) values were considered.
After docking, we have visualized the structure and the interactions using the visualization tool, Biovia Discovery Studio Visualizer (DSV) version 21.1.0 [20].
Results and Discussion
Ligand
The information related to the sinapaldehyde was retrieved from the PubChem database and the screening of it was done using Biovia Discovery Studio. The structure of the ligand was downloaded in Standard Data Format (SDF) and it was converted into PDB format for further docking purposes (Figure 1).
Figure 1: Sinapaldehyde (3D).
Receptor
See Figure 2.
Figure 2: Receptor-MCM7.
ADMET analysis
According to the Lipinski rule, there is zero violation as molecular weight was is less than 500, H-bond donors are less than 5, H-bond acceptors are less than 10 and molar refractivity is in the range of 40 to 130. As the results are in the acceptable range, we could predict that sinapaldehyde is safe for human use (Table 1-3).
Formula
C11H12O4
Molecular Weight
208.21g/mol
Num. heavy atoms
15
Num. arom. heavy atoms
6
Fraction Csp3
0.18
Num. rotatable bonds
4
Num. H-bond acceptors
4
Num. H-bond donors
1
Molar Refractivity
56.55
TPSA
55.76 Å2
Table 1: Physicochemical Properties.
GI absorption
High
BBB permeant
Yes
P-gp substrate
No
CYP1A2 inhibitor
No
CYP2C19 inhibitor
No
CYP2C9 inhibitor
No
CYP2D6 inhibitor
No
CYP3A4 inhibitor
No
Log Kp (skin permeation)
-6.58cm/s
Table 2: Pharmacokinetics.
Lipinski
Yes; 0 violation
Ghose
Yes
Veber
Yes
Egan
Yes
Muegge
Yes
Bioavailability Score
0.55
Table 3: Druglikeness.
Boiled-egg
From the boiled-egg structure, it could be predicted that the molecule (sinapaldehyde) has a high probability to permeate through the Blood-brain barrier as it is present in the yellow (yolk region) and hence it could be considered as a brain penetrant. Moreover, as it is not actively effluxed by P-glycoprotein, that is, PGP - (red dot), it could be predicted as a toxic molecule (Figure 3).
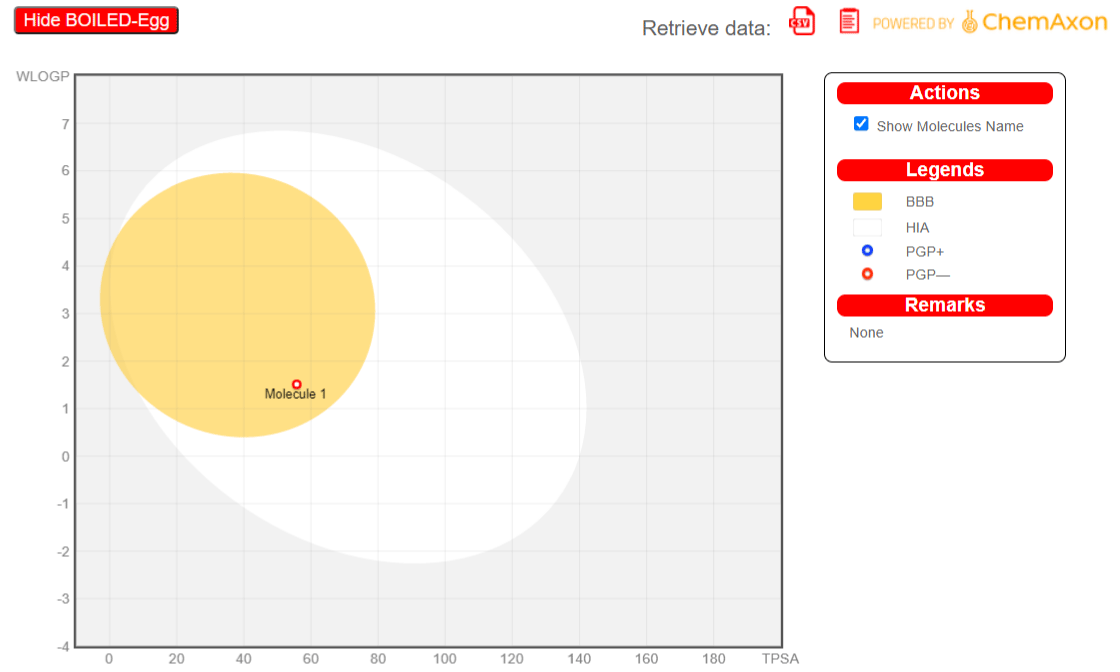
Figure 3: Hide Boiled Egg.
Molecular docking analysis
The molecular docking results show that sinapaldehyde has a good binding affinity towards MCM7. Moreover, from the Root Mean Square Deviation (RMSD), it has been found that all the transformations have been done and the protein (MCM7) is completely binding with the ligand (Table 4). By analysing the results, it could be predicted that it would be suitable for the treatment of Meier-Gorlin syndrome (Figure 4 and 5).
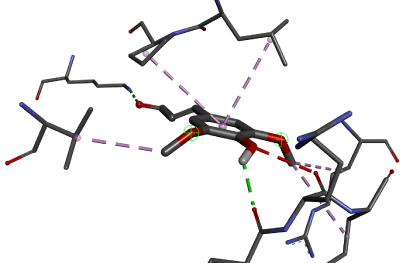
Figure 4: Ligand interactions.
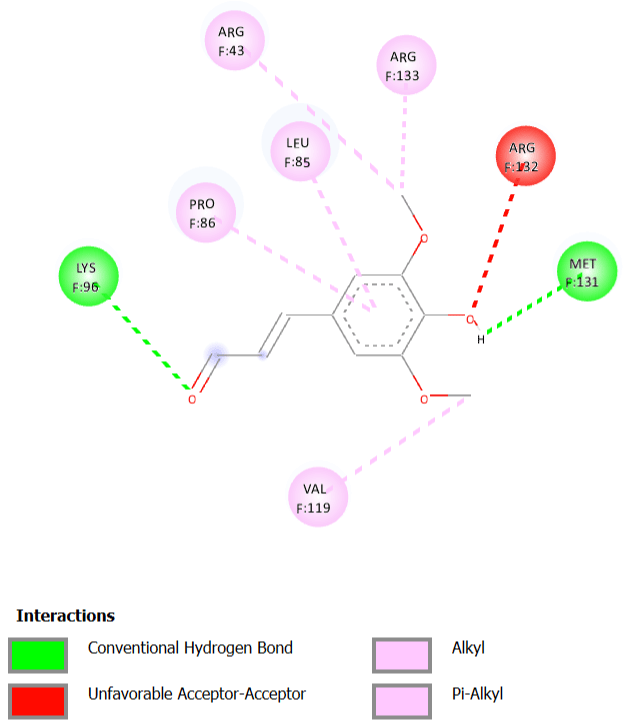
Figure 5: 2D Diagram.
Complex
Binding affinity (kcal/mol)
RMSD lower bound
RMSD upper bound
MCM7-Sinapaldehyde
-5.9
0.0
0.0
Table 4: Molecular Docking Analysis.
Conclusion
The aforementioned ligand, sinapaldehyde showed better interaction with the MCM7 receptor. By performing molecular docking of sinapaldehyde and MCM7 receptor we have found out the inhibitory activity of sinapaldehyde against the MCM7 receptor. Sinapaldehyde has the ability to inhibit the activity of MCM7 in the regular pathways with the binding affinity of -5.9kcal/mol. For understanding the therapeutic purposes we have done all the pharmacological studies followed by molecular docking. Hence, we can conclude that the ligand we have used in this study paves a way for the treatment of the Mier-Gorlin Syndrome and has also shown promising outcomes for further studies.
Declaration
Acknowledgement: The research in this publication was supported by BioNome as a part of internship program on in the field of Bioinformatics. Training on necessary online tools required for this project was given by Vaeeshnavi Buwa, BioNome, Bangalore, India.
Author’s contribution: All authors contributed equally to manuscript.
References
- de Munnik SA, Hoefsloot EH, Roukema J, et al. Meier-Gorlin syndrome. Orphanet J Rare Dis. 2015; 10: 114.
- Bongers EM, et al. Meier-Gorlin syndrome: report of eight additional cases and review. American Journal of Medical Genetics. 2001; 102: 115-124.
- Sterner, Jacqueline M., et al. “Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7”. Molecular and cellular biology. 1998; 18: 2748-2757.
- DeLano, Warren L. “Pymol: An open-source molecular graphics tool”. CCP4 Newsletter on protein crystallography. 2002; 40: 82-92.
- Seeliger Daniel, Bert L. de Groot. “Ligand docking and binding site analysis with PyMOL and Autodock/Vina”. Journal of computer-aided molecular design. 2010; 24: 417-422.
- DeLano Warren L, Sarina Bromberg. “PyMOL user’s guide”. DeLano Scientific LLC. 2004; 629.
- Dallakyan Sargis, Arthur J. Olson. “Small-molecule library screening by docking with PyRx”. Chemical biology. Humana Press, New York, NY. 2015: 243-250.
- Pawar Rutuja P, Sachin H. Rohane. “Role of Autodock vina in PyRx Molecular Docking”. Asian Journal of Research in Chemistry. 2021; 14: 132-134.
- Wang Yanli, et al. “PubChem: a public information system for analyzing bioactivities of small molecules”. Nucleic acids research. 2009; 37: W623-W633.
- Kim, Sunghwan, et al. “PubChem substance and compound databases”. Nucleic acids research. 2016; 44: D1202-D1213.
- Kim Sunghwan, et al. “PubChem 2019 update: improved access to chemical data”. Nucleic acids research. 2019; 47: D1102-D1109.
- Kiefer Florian, et al. “The SWISS-MODEL Repository and associated resources”. Nucleic acids research. 37: D387-D392.
- Schwede Torsten, et al. “SWISS-MODEL: an automated protein homologymodeling server”. Nucleic acids research. 2003; 31: 3381-3385.
- Waterhouse Andrew, et al. “SWISS-MODEL: homology modelling of protein structures and complexes”. Nucleic acids research. 2018; 46: W296-W303.
- Jenuth Jack P. “The NCBI”. Bioinformatics methods and protocols. Humana Press, Totowa, NJ. 2000: 301-312.
- Pearson William R. “Finding protein and nucleotide similarities with FASTA”. Current protocols in bioinformatics. 2016; 53: 3-9.
- Daina Antoine, Olivier Michielin, Vincent Zoete. “SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules”. Scientific reports. 2017; 7: 1-13.
- Lipinski Christopher A. “Lead-and drug-like compounds: the rule-of-five revolution”. Drug discovery today: Technologies. 2014; 1: 337-341.
- Daina Antoine, Vincent Zoete. “A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules”. Chem Med Chem. 2016; 11: 1117.
- Design LIGAND. “Pharmacophore and ligand-based design with Biovia Discovery Studio®”. 2014.