Research Article
A Proteomics. 2014;1(1): 9.
Usefulness of Urine Mass Spectrometry to Identify Women Infected with HPV 16 or Diagnosed with Higher Grades of Cervical Intraepithelial Neoplasia
Chandrika Piyathilake1*, Andres Azuero2, Roland Matthews3, Mi Kyung Kim4 and Senait Asmellash5
1Department of Nutrition Sciences, University of Alabama at Birmingham, USA
2School of Nursing, University of Alabama at Birmingham, USA
3Morehouse School of Medicine, USA
4Department of Cancer Epidemiology, National Cancer Center, Korea
5Department of Surgery, University of Alabama at Birmingham, USA
*Corresponding author: Chandrika Piyathilake, Department of Nutrition Sciences, University of Alabama at Birmingham (UAB), Wallace Tumor Institute 420D, 1824 6th Avenue South, Birmingham, AL 35294-3300, USA
Received: May 29, 2014; Accepted: August 08, 2014; Published: August 13, 2014
Abstract
Identification of women infected with human papilloma virus 16 (HPV 16) or with higher grades of cervical intraepithelial neoplasia (CIN 2+) using cost-effective and non-invasive tests will help to eliminate the worldwide burden of cervical cancer. The aim of the study was to explore the usefulness of matrix-assisted laser desorption/ionization (MALDI) time-of- flight (TOF) mass spectrometry (MS) to identify women infected with HPV 16 or diagnosed with CIN 2+ using urine samples. An additional aim was to compare several statistical and data mining techniques used to build predictive algorithms. The study used urine samples collected from 235 women diagnosed with abnormal cervical cytology. 87 and 148 women were diagnosed with CIN 2+ (cases) and ≤CIN 1 (non-cases), respectively. 126 and 109 women were tested positive or negative for HPV16, respectively. The cross-validated accuracy for detecting CIN 2+ varied from 62-73% based on the predictive technique used suggesting the usefulness of comparing different predictive modeling techniques. The positive predictive value (PPV) for detecting CIN 2+ was higher than previous studies and varied from 70% to 79%, with highest PPV noted among HPV 16 negative and African American (AA) women. Similar to CIN 2+ predictive models, the cross validated predictive accuracy for HPV 16 infections varied based on the predictive technique used, from 53% to 75%. The best PPV (75%) for HPV 16 infections was observed for AA women and the worst PPV for Caucasian American (CA) women (62%), suggesting racial differences in the usefulness of MALDI-TOF-MS based tests. The PPVs for detecting CIN 2+ or HPV16 infections were ~ 75%, a reasonably good result given the fact that non-invasively collected samples used may allow repeat testing, especially if cost-effective ELISA tests based on the discriminatory features identified in our study can be developed in the future.
\Keywords: Urine; Protein profiles; CIN
Introduction
Certain types of carcinogenic or high risk Human papilloma viruses (HR-HPVs), which are sexually transmitted, represent the most important risk factors for the development of invasive cervical cancer (ICC) as well as cervical intraepithelial neoplasia (CIN), precursor lesions for ICC [1-3]. This is unique in cancer etiology because no other human cancer has yet been shown to have a necessary cause that is so clearly identified. Information on HPV prevalence worldwide, however, is inconsistent since there is no standardized method for detecting HPV. Currently in the US, several HPV test systems exist. The US Food and Drug Administration (FDA)-approved Hybrid Capture 2 assay (HC-2) targets 13 HR-HPV genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) but does not distinguish individual HPV types. On April, 2014, the cobas HPV Test was approved by the US FDA for use as a first-line primary screening tool in women aged 25 years or older to assess risk of ICC. The test simultaneously provides pooled results for HR-HPV genotypes (HPV-31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) and individual results for HPV 16 and HPV 18.
More than 90% of cervical cancers are associated with HR-HPV DNA [4]. Studies that used improved HPV testing procedures have established HPV as a causative agent for CIN as well [5]. In the New York Cervical Disease Study, Wright et al. [6] detected HR-HPV DNA in 75% and 100% of women with low grade squamous intraepithelial lesions (LSILs) and high grade squamous intraepithelial lesions (HSILs) respectively. Eighty-five percent of women diagnosed with low grade cervical lesions in the cohorts established at the University of Alabama at Birmingham (UAB) are positive for HR-HPV based on Roche diagnostics linear array HPV genotyping test [7]. Results from this cohort also showed that among women diagnosed with LSIL and HSIL where a large majority are positive for HR-HPV, only 23% and 50% respectively have biopsy-confirmed higher grades of CIN (CIN 2+). These observations demonstrate that the presence of any HR-HPV genetic material is not by itself indicative of cervical disease. Therefore, novel markers with higher specificity for the presence of CIN 2+ among HR-HPV positive are needed and this will improve cervical cancer screening and reduce the cost associated with patient care.
Infection with HPV 16 is the biggest causative agent of cervical cancer and it is the most prevalent HR-HPV in the USA. Campion et al. [8] reported that 58% of HPV 16-positive women who had mild cervical atypia progressed to CIN 3 within 2 years. The Centers for Disease Control (CDC) estimates that 20 million people in the US are infected with HPV 16 and that every year there are about 5.5 million new infections. Results from our studies [7] demonstrate that the prevalence of HPV 16 in our population can vary from 21% to 55% depending on the grade of cervical lesions.
Because there is no cure for infection with HPVs, prevention and control of these infections could be used for primary cancer prevention, possibly saving thousands of lives. Because HPV 16 is the most prevalent HPV genotype associated with CIN and ICC, its detection and control would clearly offer a cost effective long-term strategy to reduce the cervical cancer burden. HPV vaccine development holds great promise for reducing the incidence of ICC, and the addition of a vaccine against HPV 16 is projected to be a cost-effective use of health care resources [9]. However, a type-specific HPV vaccine may reduce but not eliminate the risk of ICC [10]. Therefore, cervical cancer screening recommendations are unlikely to change for females who receive the HPV vaccine. Because of inadequate data on its long-term effectiveness, the impact of type-specific vaccines for other HR-HPVs, and duration of immunity, it is unlikely that routine screening programs or other preventive measures will be replaced by HPV vaccines in the near future. Therefore, at this point, a vaccine approach may not reduce the health care cost associated with prevention efforts for cervical cancer. Because of this, development of cost effective tests that can be used to identify women at risk is more important than ever. Tests that may identify women infected with HPV 16 are of high importance because of its higher carcinogenicity. As discussed below, such tests are also important for providing HPV vaccines to the most appropriate individuals.
Ideally, HPV prophylactic vaccine should be administered before the onset of sexual activity and females who have not been infected with any vaccine HPV type would receive the full benefit of vaccination. Females who are already sexually active but not infected with vaccine HPV type would still get protection from the vaccine but currently, there is no cost-effective test available for clinical use to determine whether a female has had any or all of the four HPV types in the currently available vaccine (HPV 6, 11, 16, and 18). Therefore, development of cost-effective tests which are able to detect HPV infections, especially, HPV 16 will not only be useful for routine screening but also for identifying women who will benefit from HPV vaccines even after the onset of sexual activity.
Protein biomarkers have a great potential in elucidating the biology of disease progression. Biomarker discovery efforts have shown that proteins such as the cyclin-dependent kinase inhibitor p16 are differentially expressed in normal versus cervical cancer cells. The potential candidates described so far require in situ hybridization or immune histochemistry of tissue samples. Ideally, we would like to find biomarkers that are easily detected in non-invasively collected samples, such as urine, using an assay that could be adapted for high throughput clinical applications. The merits of MALDI MS profiling and capillary electrophoresis coupled with electrospray ionization (ESI) MS has been recently reviewed by Albalat et al. [12]. While each platform offers distinct advantages, MALDI MS, is a particularly rapid and simple technique for analyzing complex biospecimens such as urine, serum and plasma [13].
The aim of this work was to explore the usefulness of urine mass spectrometry to identify women infected with HPV 16 or diagnosed with CIN 2+. Since there is no standard way of analyzing MALDI MS profiling data, an additional aim of the study was to use several statistical and data mining techniques to compare the accuracy of results generated by different predictive modeling techniques.
Materials and Methods
Patient population
The study was based on the analysis of urine samples collected from 235 women referred for colposcopy because of abnormal cervical cytology. All women were diagnosed with abnormal pap and were enrolled in a prospective follow-up study funded by the National Cancer Institute (R01 CA105448, Prognostic Significance of DNA & Histone Methylation). The parent study has been described in a previous publication [7]. The study protocol and procedures were approved by the UAB Institutional Review Board. Among the 235 women, 87 were diagnosed with CIN 2+ (cases) and 148 were diagnosed with ≤CIN 1 (non-cases). The average age among the women was 24.5years (SD=5.1, Range=19-48); 58% (n=136) were African American (AA) and 42% (n=99) identified themselves as Caucasian American (CA). Additional participant characteristics by case status are shown in Table 1. Parity was significantly higher among cases compared to controls as previously reported for this study population [7]. Urine peptide/protein profiles from this sample were used to predict CIN 2+ status. Because 126 women out of the 235 tested positive for HPV16, the urine peptide/protein profiles were also used to predict HPV16 infection status.
Characteristic
≤CIN1 (N=148)
CIN2+ (N=87)
P
Age in years, mean (SD)
24.4 (5.2)
24.8 (4.9)
0.51
Body Mass Index (BMI), kg/m2, mean (SD)
28.3 (8.7)
27.2 (7.2)
0.34
Parity, n (%)
0.03
0
53 (35.8)
20 (23.0)
1
53 (35.8)
26 (29.9)
2
30 (20.3)
29 (33.3)
3
12 (8.1)
12 (13.8)
African American, n (%)
89 (60.1)
47 (54.0)
0.36
HPV16 positive, n (%)
79 (53.4)
47 (54.0)
0.92
Table 1: Participant characteristics by the case status.
Testing for HPVs
DNA was extracted from cervical cells using the QIA amp Mini Elute Media Kit (Qiagen, Inc., Valencia, CA) following the manufacturer's instruction for HPV genotyping test. HPV genotyping test (Linear array, Roche diagnostics) was performed according to the manufacturer's instructions by a research associate trained by personnel from Roche Diagnostics. Briefly, target DNA amplified by Polymerase Chain Reaction (PCR) utilized the PGMY09/11 L1 consensus primer system and included co-amplification of a human cellular target, β-globin, as an internal control. Detection and HPV genotyping were achieved using a linear array HPV genotyping test which included probes to genotype for 37 anogenital HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73 (MM9), 81, 82 (MM4), 83 (MM7), 84 (MM8), IS39, and CP6108).
Generation of Protein Profiles
Mass Spectrometry
Sample preparation: Urine samples collected from patients were stored at -80°C until analysis. A high affinity solid core lipophilic extraction resin was used to desalt and prepare the urine sample MALDI MS analysis. Bondapak C18 125 A, 37-55 μm resin (Waters, Milford, MA, USA) was packed into 96 well 0.45 μm Unifilter plates (Whatman, Florham Park, NJ, USA). Packed resins were activated with 80% acetonitrile: 20% water. Urine samples were thawed, acidified by adding Tri Fluoro Acetic acid (TFA) to a final concentration of 1% TFA v/v, and mixed with the activated C18 resins. The unbound urine fraction was removed by centrifugation of the 96 well plates for 5 min at 1500 g. The resin was washed twice with 200 μl of 1% TFA per well, and the bound low molecular weight proteins and peptides were eluted with 100 μl of 70% CH3CN:30% water with 0.1% TFA. Eluants were mixed with an equal volume of matrix consisting of 20 mg/ml sinapinic acid (Fluka, St. Louis, MO, USA) in 50:50 CH3CN: water with 0.1% TFA and spotted onto a MALDI target plate for MALDI-TOF analysis.
Mass profiling
Mass spectra were acquired in linear positive ion mode using an Ultra flex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics). Instrument settings optimized for mass range m/z 2-20 kDa were used: ion source 1 = 25.0 kV, ion source 2 = 23.45 kV, pulsed ion extraction time = 15 ns. Mass calibration was carried out externally using a mixture of standards consisting of insulin, cytochrome C, myoglobin and ubiquitin (Bruker Daltonics, Bremen, Germany).
Data analysis
Data obtained from the mass spectra was exported as text files for preprocessing and further analysis. Spectral preprocessing included baseline correction, noise estimation, background estimation, normalization (using total ion current) and finally peak picking. The preprocessing step resulted in 171 features in the mass-to-charge (m/z) range of 2-20 kDa.
Two sets of analyses were conducted for this study. In the first set, the urine peptide/protein profiles were utilized to predict CIN 2+ status. In the second set of analyses, the urine protein profiles were utilized again but to predict HPV16 infection status. In addition to the 171 m/z features from the MALDI MS profiles, other predictors included age, body mass index (BMI), parity, and race (AA vs. CA).
Within each set of analyses, several statistical and data mining techniques were applied to the peak data in order to obtain prediction algorithms. All analyses were conducted using the R statistical software [14]. The predictive analytical techniques are briefly described below.
Weighted k-nearest neighbors
The classical k-nearest neighbor technique is a non-parametric classification method where a new observation is compared to the k closest available observations (with respect to some covariates and a distance measure), and is then assigned to the majority class among these closest k observations. The weighted k-nearest neighbor technique [15] is an extension of the classical method that weighs the majority class assignment for the new observation by the distance between the new observation and each of the k closest available observations, with closer observations having more weight in the class assignment. The weight can be determined by different mathematical functions. For this study, we used the implementation of the technique in the R package kknn [16] Using leave-one-out cross-validation, the kknn algorithm searches for the optimal number k and weight function (from nine choices). The default Euclidean distance was used as distance metric. Because the algorithm may perform poorly in the presence of irrelevant covariates, a first step consisted of screening the predictors to be used as covariates with Wilcoxon tests or Kolmogorov-Smirnov two-sample tests (p-value <.2), and then in a second step the weighted k-nearest neighbor algorithm was applied. Leave-one-out cross-validation was conducted for the two-step procedure.
Nearest shrunken centroids
The nearest centroid method is a classification technique similar to linear discriminant analysis. A new observation is assigned to the class for which the multivariate distance between the observation's covariate values and the class centroid (vector of means) is shortest. The nearest shrunken centroids method [17] is an extension of the original technique developed for high dimensional problems and that includes built-in covariate selection using cross-validation. For this study we used the implementation of the technique in the R package pamr [18]. Using 10-fold cross-validation the pamr algorithm searches for the value of a tuning parameter involved in the selection of predictors to include in the centroid and distance computations. Classification results from the 10-fold cross-validation are reported.
Least angle regression (LAR)
LAR [19] is a forward stepwise linear regression technique developed for high dimensional problems. To be used in binary classification, the outcome is coded as 1 or -1 and a cutoff value of zero is applied to the prediction [20]. For this study we used the implementation of the technique in the R package lars [21]. Using 10-fold cross-validation, the lars algorithm searches for the optimal number of predictors to be included in the regression model. Results from the 10-fold cross-validation are reported. Three models were implemented: (1) a model with features and additional predictors as main effects; (2) a model with main, quadratic, and cubic effects for all predictors; and (3), a model with main, quadratic, cubic effects for each predictor, and all two-way interaction terms between predictors.
Logistic regression with elastic net
The term elastic net [18] refers to a forward stepwise model selection technique in generalized linear models such as logistic regression, developed for high dimensional problems. For this study we used the implementation of the technique in the R package glmnet [22]. Using 10-fold cross-validation the glmnet algorithm searches for the value of a tuning parameter involved in the selection of predictors to include in the model. Cutoff predicted probability for classification was at 0.5. Classification results from the 10-fold cross-validation are reported. Three models were implemented: (1) a model with features and additional predictors as main effects; (2) a model with main, quadratic, and cubic effects for all predictors; and (3), a model with main, quadratic, cubic effects for each predictor, and all two-way interaction terms between predictors.
Principal component regression (PCR) and Partial least squares regression (PLSR)
Given a set of correlated variables, the principal components are a smaller set of non-correlated synthetic variables computed as linear combinations of the original variables, and that capture most of the information contained in the original correlated variables [23]. PCR is a linear regression where the covariates are the principal components of the explanatory variables originally measured. To be used in binary classification, the outcome is coded as 1 or -1 and a cutoff value of zero is applied to the prediction. PLSR is a modification of PCR, in that the outcome is also included in the computation of the principal component scores [24]. Typically, PCR and PLSR achieve similar prediction accuracies, but PLSR needs fewer components than PCR. For this study we used the implementation of the techniques in the R package pls [25]. Using Leave-one-out cross-validation, the pls algorithm searches for the optimal number of components extracted from the features and additional predictors to be included in a regression model as covariates (main effects only). Results of leave-one-out cross-validation are reported.
Classification trees
Classification tree models are created by recursive partitioning, i.e. splitting the predictor space recursively into disjoint regions and then assigning the class of each resulting cell to the majority class among the observations included in that cell. The resulting models can be represented as binary trees. Two common algorithms were used in this study. The first is the non-parametric CART (Classification and Regression Tree) algorithm [26] that builds the tree model in two stages: first, the single variable is found which best (with regards to the target classification) splits the observations into two groups. The data are separated, and then this process is applied separately to each sub-group, and so on recursively until the subgroups either reach a minimum size or until no improvement can be made. The second stage consists of using cross-validation to "prune" or trim back the initial full tree to a size that provides the best cross-validated accuracy. For this study we used the implementation of the technique in the R package rpart [27]. Results of leave-one-out cross-validation are presented. A second algorithm used in this study builds conditional inference tree models [28]. As opposed to CART, this algorithm builds the tree model in one stage by establishing the splits with significance tests (adjusted for multiple testing), so that the stopping criterion for splitting is statistical (based on p-values), and no pruning is needed. We used the implementation of the algorithm provided in the R package party [29]. Leave-one-out cross-validation results are presented.
Ensembles of classification trees
The term ensemble refers to a method to generate many classification models and combine their results. The classification for a new observation is produced by the ensemble as a combination of the predicted classifications from each of its individual models. Using CART tree models, two common ensemble methods were applied in this study. The first method is Random Forests [30]. It consists of generating a number n of bootstrap re hyphen samples from the original dataset, and for each resample building an untrimmed tree model; however for each split in a tree, only a small random set of predictors are available for consideration. The resulting ensemble is composed of n trees, and the classification for a new observation is given by the majority class among each of the individual tree predictions. An unbiased estimate of accuracy is given by classifying each observation with only those trees corresponding to bootstrap re hyphen samples in which the observation did not appear. In Random Forest implementations, this estimate of accuracy is referred to as 'out-of-bag' accuracy, and is equivalent to a cross-validated estimate of accuracy [18]. We used the implementation of the technique in the R package random Forest [28], with n=500 re hyphen samples. Out-of-bag classification results are presented. A second ensemble method used in this study is Adaptive Boosting [31]. This algorithm associates weights to each observation, and builds a tree ensemble sequentially. After each new individual tree model is built, the observation weights are updated by increasing (boosting) the weights of misclassified observations, so that in the next individual tree model in the sequence, these misclassified observations are given more importance. The final prediction is a combination of the predictions from each individual tree model in the sequence. We used the implementation of the technique in the R package ada [32], with a sequence of up to 150 individual trees. Results from 10-fold cross-validation are presented.
Because an ensemble model is a collection of hundreds of individual models, loss of interpretability is a drawback. The algorithms, however, compute measures of relative predictor importance (for instance by counting the most frequent predictors in the ensemble) that allow ranking of predictors by their usefulness.
Artificial neural networks (ANNs)
An ANN is an extension of a generalized linear model [18,33] that can be used to model complex linear and non-linear relationships between a set of covariates and an outcome. It is inspired by the functional aspects of biological neurons, with a set of inputs, a set of intermediary 'neurons' or nodes (referred to as a 'hidden layer' of neurons) and a set of outputs. A model with one set of intermediary nodes or neurons (one 'hidden layer') works as a two-stage regression. In the first stage, the algorithm computes values for the intermediary neurons or nodes as linear combinations of the inputs (i.e. the covariates or predictors). In the second stage, the values of the intermediary neurons form a second linear combination to then model the output (i.e. the outcome variable). Because the algorithm does not have built-in predictor selection, and may perform poorly in the presence of irrelevant covariates, a first step consisted of screening the features and additional predictors to be used as inputs or covariates with Wilcoxon tests (p-value<.2), and then in a second step the ANN models were fitted. We used the implementation of the technique in the R package neuralnet [34]. We implemented models with one hidden layer fitted with up to 20 neurons. Results from 10- fold cross-validation, conducted for the two-steps (peak selection and model fitting), are reported.
Cross-validated accuracy results were complemented by computation of measures of sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) for the whole sample as well as for the following subgroups: 1) race groups; 2) HPV16 infection (for prediction of CIN 2+); and 3) case status (for prediction of HPV16 infection).
Results
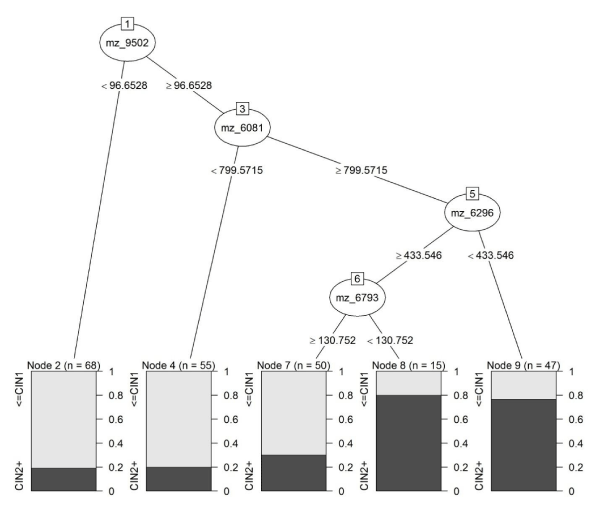
Cross-validated accuracy results for the two sets of analyses are presented in Table 2. For the first set of analyses (predicting CIN 2+) a classification tree with the CART algorithm provided the best cross-validated prediction (73% accuracy). Cross-validated sensitivity, specificity and PPV were estimated at 85%, 51% and 75%, respectively (Table 3). Accuracy of the prediction among HPV16 negative samples was slightly higher than among HPV16 positive samples (76% vs. 70%), however the difference was not statistically significant (p=.296). Accuracy among AAs was slightly higher than among CAs (75% vs. 70%; p=.394). The CART model used the following four features: m/ z=6081, m/z=6296, m/z=6793, and m/z=9502. The model is shown in Figure 1.
Figure 1 : Classification tree model for CIN 2+ status using intensities for features m/z=6081, m/z=6296, m/z=6793, and m/z=9502 as predictors.
Cross-validated Prediction Accuracy (%)
Predictive technique
CIN2+
HPV16 infection
N=235 (87 CIN2+)
N=235 (126 positive)
K-nearest neighbors
Featuresscreened with Wilcoxon test
62
65
Features screened with K-S test
62
66
Shrunken centroids
65
69
Least Angle Regression
Main effects (ME)
63
65
ME+Quadratic and Cubic effects (QCE)
63
68
ME+QCE+Two-way interactions
63
53
Logistic regression with elastic net
Main effects (ME)
63
70
ME+Quadratic and Cubic effects (QCE)
63
66
ME+QCE+Two-way interactions
63
75
Partial least squares regression
63
68
Principal Components Regression
63
71
Classification trees and tree ensembles
CART
73
56
Conditional
63
61
Random Forest with CART
63
70
Boosting with CART
63
70
Artificial Neural Networks
Features screened with Wilcoxon test
63
60
Table 2: Cross-validated accuracy results from predictive analytical techniques.
Subgroups
N Target/NTotal
Accuracy (%)
Sensitivity (%)
Specificity (%)
*PPV (%)
*NPV (%)
All
87/235
73
85
51
75
67
HPV16
Positive
47/126
70
86
42
71
64
Negative
40/109
76
84
61
79
69
Race
African American
47/136
75
84
57
79
66
Caucasian American
40/99
70
87
44
70
69
Table 3: Cross-validated prediction results for case status by subgroups.
In the second set of analyses (predicting HPV16 infection) a complex logistic regression model consisting of a cubic term and 32 two-way interactions between predictors provided the best cross-validated prediction with 75% accuracy (Table 2). Cross-validated sensitivity and specificity were estimated at 77% and 74%, respectively (Table 4). Accuracy of the prediction among ≤CIN 1 samples was slightly higher than among CIN 2+ samples (76% vs. 74%; p=.731). Accuracy among AAs was slightly higher than among CAs (76% vs. 73%, p=.523). With all 235 samples, the following 37 features were used in the logistic model: m/z=2111, m/z=2152, m/z=2208, m/ z=2436, m/z=2477, m/z=2569, m/z=2755, m/z=2878, m/z=2943, m/ z=3255, m/z=3444, m/z=4243, m/z=4483, m/z=4833, m/z=4922, m/ z=5252, m/z=6066, m/z=6338, m/z=6404, m/z=6435, m/z=6550, m/ z=7206, m/z=7891, m/z=8020, m/z=8051, m/z=8188, m/z=8301, m/ z=8938, m/z=9077, m/z=9192, m/z=9219, m/z=9502, m/z=9866, m/ z=10162, m/z=12439, m/z=20191, m/z=20351. Additional predictors in the model included age, BMI and race. The model is shown in Table 5. Because of the model's complexity, traditional interpretation of the individual parameters is unfeasible.
Subgroups
N target/ N total
Accuracy (%)
Sensitivity (%)
Specificity (%)
*PPV (%)
*NPV (%)
All
126/235
75
77
74
73
78
Case status
CIN2+
47/87
74
73
74
71
76
≤CIN1
79/148
76
79
73
73
79
Race
African American
58/136
76
87
62
75
78
Caucasian American
68/99
73
53
84
62
78
Table 4: Cross-validated prediction results for HPV16 infection by subgroups.
Model Parameter
Coefficient Estimate
Intercept
-1.18E+00
(m/z=2436)3
-6.80E-01
BMI x Age
-2.37E-04
AA x m/z=5252
-4.55E-04
m/z=2111 x m/z=6338
5.86E-10
m/z=2152 x m/z=20351
7.44E-07
m/z=2208 x m/z=2943
-2.93E-08
m/z=2477 x m/z=6338
2.04E-07
m/z=2477 x m/z=20191
1.33E-06
m/z=2569 x m/z=4922
-4.89E-08
m/z=2755 x m/z=8301
-1.48E-08
m/z=2878 x m/z=9866
7.71E-08
m/z=2943 x m/z=9502
-6.24E-08
m/z=3255 x m/z=6066
1.37E-07
m/z=3255 x m/z=6435
4.39E-07
m/z=3255 x m/z=7891
4.56E-07
m/z=3255 x m/z=8020
2.08E-07
m/z=3255 x m/z=20351
1.12E-06
m/z=3444 x m/z=4483
7.29E-09
m/z=4243 x m/z=6338
2.62E-07
m/z=4833 x m/z=8188
-2.76E-07
m/z=6066 x m/z=6338
5.65E-08
m/z=6066 x m/z=7206
5.40E-07
m/z=6066 x m/z=8020
7.32E-08
m/z=6066 x m/z=8051
5.63E-08
m/z=6338 x m/z=8051
1.50E-07
m/z=6338 x m/z=8938
1.70E-07
m/z=6338 x m/z=9192
6.96E-08
m/z=6338 x m/z=20351
7.15E-07
m/z=6404 x m/z=8301
-2.34E-07
m/z=6435 x m/z=20351
2.55E-06
m/z=6550 x m/z=9077
4.17E-08
m/z=9077 x m/z=10162
2.52E-07
m/z=9219 x m/z=12439
3.08E-06
Table 5: Logistic model for prediction of HPV16 infection status.
Descriptive statistics of intensity (measured in total ion current) for the features included in the final CART model (predicting CIN 2+) an in the logistic model (predicting HPV16 infection) are shown in Table 6.
Listed subject with IPF
Donor listing
Offer A
Offer B
Offer C
Age
65 years
12-60 years
25 years
25 years
32 years
Gender
Male
Either
Female
Male
Female
Height (cm)
170
147 - 170
147
170
175
pTLC (liter)
6.54
3.98- 6.54
3.98
6.54
5.76
pTLC ratio
0.61
1.00
0.88
Table 6: Descriptive statistics of intensity (measured in total ion current) for the features included in the final CART model, predicting CIN2+, and in the logistic model, predicting HPV16 infection.
Discussion
Even though HR-HPV is a necessary factor for the development of CIN and ICC, being positive for HR-HPVs does not invariably lead to such lesions [2]. Several studies have shown that the detection of HR-HPVs provides high sensitivity, but has lower specificity for the identification of CIN 2+ lesions in screening populations in many countries [35,36]. A recent study which compared seven HPV tests and p16 INK4a cytology in a high risk population demonstrated that the PPV for detecting high grade CIN varied from ~37% to 55% and 49% respectively [37]. These tests, therefore, would always require follow-up tests including referrals to invasive colposcopic procedures to confirm diagnosis and avoid patient anxiety. Numerous attempts have been made to identify protein biomarkers with higher PPV for detecting high grade CIN. Most studies focused on proteins involved in cell cycle regulation, signal transduction, DNA replication or cellular proliferation [38,39]. The PPV of most of these tests for detecting high grade CIN also has been ~50% [40].
An important application of MALDI-TOF MS is the simultaneous analysis of multiple proteins to establish "fingerprint" profiles that discriminate disease from non-disease. This is an important approach, since no single biomarker or protein alone may be sufficient for detecting high grade CIN with high PPV. If these types of testing can be performed using non-invasive and easy to collect samples such as urine, the clinical utility of such tests is likely to be enormous. Our results demonstrated that the accuracy for detecting CIN 2+ based on urine protein profiles varied from 62-73% based on the predictive technique used suggesting the usefulness of utilizing and comparing different predictive modeling techniques. We also observed that the PPV for detecting CIN 2+ is higher than previous studies and varied from 70% to 79%, with highest PPV noted among HPV 16 negative women and AA women. For modeling the relationship between CIN 2+ and urine protein profiles, the non-parametric CART model that provided the best prediction appeared to be somewhat simple, utilizing only 4features.
While most HPV infections are asymptomatic and transient, HPV is of clinical and public health importance because persistent infection with certain HR-HPV types can lead to CIN 2+ or ICC. Repetition of the cytology/colposcopy/biopsy-based screening for HPV associated changes in the cervix has led to substantial decreases in cervical cancer rates in countries that have sufficient resources to sustain a high-quality, organized screening programs [41]. All these approaches have limitations in terms of sensitivity and specificity [42] and higher health care costs as a result of those issues. HR-HPV testing as an adjunct to cytology in primary cervical cancer screening is now accepted [43], but whether this type of testing implemented into an existing public-health screening program can result in an increase in the program effectiveness is somewhat controversial. These methods are even less useful in developing countries, which currently carry the greatest burden of cervical cancer incidence and mortality, due to the expense and expertise required for the gynecologic exams and sample collection for cytological and HPV tests. Because HPV testing that distinguishes HPV16 and HPV18 from other HR-HPV types has been shown to identify women at the greatest risk of CIN 3+, this type of testing is more likely to be clinically useful than a pooled HPV test [44]. Even though only a fraction of women infected with HPV16 develop CIN 2+, these lesions have the highest rate of progression to ICC [45]. Further, the recurrence rate of CIN 2+ after a loop electrosurgical excision procedure (LEEP) was shown to be significantly higher among those who were tested positive for HPV 16 before and after the procedure [46]. Therefore, identification of this fraction of women and treatment of their lesions and closer follow-up after treatment are important unmet medical needs in the current management protocols. Further, as discussed in the introduction, such tests are also important for providing HPV vaccines to the most appropriate individuals.
Currently available tests do not have adequate specificity for identifying women with HPV 16-associated CIN 2+. In the patient sample used in this study, 37% (n=47) of the 126 women infected with HPV 16 were diagnosed with CIN grades higher than 2 (CIN 2+). Identification, treatment and closer follow-up of these women would offer a cost-effective strategy to reduce the cervical cancer burden. A meta-analysis showed that detecting any HR-HPV by the Hybrid Capture 2 test among women with abnormal pap demonstrated 97.2% sensitivity for detecting CIN 2+ and 97.1% sensitivity for detecting CIN 3+. This analysis also demonstrated a pooled specificity of 30.6% and 26.1% when the outcome was CIN 2+ and CIN 3+ respectively [47]. A recent study demonstrated that the sensitivity of the HPV 16/18 genotyping test for detection of CIN 2+ was > 93% while the specificity of the test for detection of CIN 2+ and CIN 3+ was 44.2% and 43%, respectively [48]. Two protein features identified in serum in one of our previous studies demonstrated higher specificity for identifying CIN 2+ among HPV 16 positive women than these published studies [49].
The results from the current exploratory study suggested that the relationship between HPV16 infection status and urine protein profiles is complex, and might only be modeled by complex algorithms that include non-linear effects and interactions among a fairly large number of features. Similar to CIN 2+ predictive models, the cross validated predictive accuracy for HPV 16 infections varied based on the predictive technique used, from 53% to 75%. The best PPV (75%) for HPV 16 infections was observed for AAs and the worst PPV for CAs (62%), suggesting racial differences in usefulness of MALDI-TOF-MS based tests. To our knowledge, this is the first study to evaluate the usefulness of urine protein profiles for identifying women infected with HPV 16. PPV was similar among cases and controls suggesting that urine profiles are useful in identifying HPV infections regardless of lesion status.
We demonstrated that the PPVs for detecting CIN 2+ or HPV16 infections are ~ 75%, a reasonably good result given the fact that non-invasively collected samples used may allow repeat testing, especially if cost-effective ELISA tests based on protein features identified in our study can be developed in the future. However, the observed PPVs may not be high enough to be used in triage of patients, especially in populations exposed to HPV 16/18 prophylactic vaccines where the rate of HPV 16 infections and CIN 2+ are likely to be reduced resulting in further lowering of the PPV value of these tests. Therefore, continuation of biomarker research with other non-invasively collected samples are needed to discover and validate biomarkers with higher PPV to identify and treat only those women truly at high risk for developing CIN 2+ or ICC in the future.
Acknowledgement
This work was supported by U54 CA118948 and R01 CA105448 funded by the National Cancer Institute and a Fulbright Senior Scholar Research Grant awarded to Chandrika J Piyathilake, administered via the Korean-American Educational Commission (2012, Chungbuk National University).
References
- Franco EL, Rohan TE, Villa LL. Epidemiologic evidence and human papillomavirus infection as a necessary cause of cervical cancer. J Natl Cancer Inst. 1999; 91: 506-511.
- JM Walboomers, MV Jacobs, MM Manos, FX Bosch, JA Kummer, KV Shah, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999; 189: 12-19.
- Human papillomaviruses. IARC Monographs on the evaluation of carcinogenic risks to humans. Lyon, France: International Agency for Research on Cancer. 1995; 64.
- FX Bosch, MM Manos, N Munoz, M Sherman, AM Jansen, J Peto, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group, JNatlCancerInst. 1995; 87: 796-802.
- MH Schiffman, HM Bauer, RN Hoover, AG Glass, DM Cadell, BB Rush, et al. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J NatlCancerInst. 1993; 85: 958-964.
- TC Wright, TV Ellerbrock, MA Chiasson, NV anDevanter, XW Sun. Cervical intraepithelial neoplasia in women infected with human immunodeficiency virus: prevalence, risk factors, and validity of Papanicolaou smears. New York Cervical Disease Study. Obstetrics & Gynecology. 1994; 84: 591-597.
- Piyathilake CJ, Macaluso M, Alvarez RD, Bell WC, Heimburger DC, Partridge EE, et al. Lower risk of cervical intraepithelial neoplasia in women with high plasma folate and sufficient vitamin B12 in the post-folic acid fortification era. Cancer Prev Res (Phila). 2009; 2: 658-664.
- MJ Campion, DJ McCance, J Cuzick, A Singer. Progressive potential of mild cervical atypia: prospective cytological, colposcopic, and virological study. Lancet2. 1986; 328: 237-240.
- Goldie SJ, Kohli M, Grima D, Weinstein MC, Wright TC, Bosch FX, et al. Projected clinical benefits and cost-effectiveness of a human papillomavirus 16/18 vaccine. J Natl Cancer Inst. 2004; 96: 604-615.
- Hughes JP, Garnett GP, Koutsky L. The theoretical population-level impact of a prophylactic human papilloma virus vaccine. Epidemiology. 2002; 13: 631-639.
- Kulasingam SL, Myers ER. Potential health and economic impact of adding a human papillomavirus vaccine to screening programs. JAMA. 2003; 290: 781-789.
- Albalat A, Husi H, Stalmach A, Schanstra JP, Mischak H. Classical MALDI-MS versus CE-based ESI-MS proteomic profiling in urine for clinical applications. Bioanalysis. 2014; 6: 247-266.
- Dudley E. MALDI Profiling and Applications in Medicine. Adv Exp Med Biol. 2014; 806: 33-58.
- Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 2007; 13: 497-512.
- K Hechenbichler, K Schliep. Weighted k-Nearest-Neighbor Techniques and Ordinal Classification, Discussion Paper 399, SFB 386, Ludwig-Maximilians University Munich. 2004.
- K Schliep, K Hechenbichler. kknn: Weighted k-Nearest Neighbors. R package version 1.0-8. 2010.
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002; 99: 6567-6572.
- T Hastie, R Tibshirani, B Narasimhan, G Chu. pamr: Pam: prediction analysis for microarrays. R package version 1.51. 2013.
- B Efron, T Hastie, I Johnstone, R Tibshirani. Least angle regression (with discussion), Annals of Statistics. 2004; 32: 407-499.
- T Hastie, R Tibshirani, J Friedman. The Elements of Statistical Learning, 2nd ed. New York: Springer-Verlag. 2009.
- T Hastie, B Efron. lars: Least Angle Regression, Lasso and Forward Stagewise. R package version 0.9-8. 2011.
- Friedman J1, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010; 33: 1-22.
- B. Manly. Multivariate statistical methods: a primer, 3rd ed. Boca Raton, FL: Chapman & Hall/CRC. 2005; 75-76.
- R Wehrens, B Mevik. The pls Package: Principal Components and Partial Least Squares Regression in R. Journal of Statistical Software. 2007; 18: 1-24.
- R Wehrens, B Mevik. pls: Partial Least Squares Regression (PLSR) and Principal Component Regression (PCR). R package version. 2.1-0. 2010.
- T Therneau, E Atkinson. Technical Report Series No. 61, An introduction to Recursive Partitioning Using the RPART Routines. Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, Minnesota. 1997.
- T Therneau, E Atkinson, B Ripley. rpart: Recursive Partitioning. R package version. 2011; 3: 1-50.
- T Hothorn, K Hornik, A Zeileis. Unbiased Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical Statistics. 2006; 15: 651-674.
- T. Hothorn, K. Hornik and A. Zeileis. party: A Laboratory for Recursive Partitioning. R package version 0. 2009; 9-996.
- A Liaw, M Weiner. Classification and Regression by random Forest. R News 2. 2002; 18-22.
- M Culp, K Johnson, G Michailidisada. An R package for stochastic boosting. Journal of Statistical Software. 2006; 17: 1-27.
- M Culp, K Johnson, G Michailidis. ada: an R package for stochastic boosting. R package version 2.0-2. 2010.
- F Gunther, S Fritsch. Neuralnet: Training of Neural Networks. R Journal. 2010; 2: 30-38.
- S Fritsch, F Guenther, M Suling. Neuralnet: Training of neural networks. R package version 1.31. 2010.
- J Cuzick, A Szarewski, H Cubie, G Hulman, H Kitchener, D Luesley, et al. Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet. 2003; 362: 1871-1876.
- Kotaniemi-Talonen L, Anttila A, Malila N, Tarkkanen J, Laurila P, Hakama M, et al. Screening with a primary human papillomavirus test does not increase detection of cervical cancer and intraepithelial neoplasia 3. Eur J Cancer. 2008; 44: 565-571.
- A Szarewski, D Mesher, L Cadman, J Austin, L Ashdown-Barr, L Ho, et al. Comparison of seven tests for high-grade cervical intraepithelial neoplasia in women with abnormal smears: the Predictors 2 study. J ClinMicrobiol. 2012; 50: 1867-1873.
- Malinowski DP. Multiple biomarkers in molecular oncology. I. Molecular diagnostics applications in cervical cancer detection. Expert Rev Mol Diagn. 2007; 7: 117-131.
- Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007; 23: 315-330.
- Dunton CJ, van Hoeven KH, Kovatich AJ, Oliver RE, Scacheri RQ, Cater JR, et al. Ki-67 antigen staining as an adjunct to identifying cervical intraepithelial neoplasia. Gynecol Oncol. 1997; 64: 451-455.
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55: 74-108.
- MH Stoler, M Schiffman. Atypical Squamous Cells of Undetermined Significance-Low-grade Squamous Intraepithelial Lesion Triage Study (ALTS) Group, Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study, JAMA. 2001; 285: 1500-1505.
- Wright TC Jr, Schiffman M, Solomon D, Cox JT, Garcia F, Goldie S, et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol. 2004; 103: 304-309.
- Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005; 97: 1072-1079.
- K Matsumoto, A Oki, R Furuta, H Maeda, T Yasugi, N Takatsuka, et al. Japan HPV and Cervical Cancer (JHACC) Study Group, Predicting the progression of cervical precursor lesions by human papillomavirus genotyping: A prospective cohort study, Int J Cancer. 2011; 128: 2898-2910.
- WD Kang, MJ Oh, SM Kim, JH Nam, CS Park, HS Choi. Significance of human papillomavirus genotyping with high-grade cervical intraepithelial neoplasia treated by a loop electrosurgical excision procedure, Am J Obstet Gynecol. 2010; 203: 72.e1-6.
- J Cuzick, M Arbyn, R Sankaranarayanan, Tsu V, Ronco G, MH Mayrand, et al. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008; 26: K29-K41.
- Einstein MH, Martens MG, Garcia FA, Ferris DG, Mitchell AL, Day SP, et al. Clinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytology. Gynecol Oncol. 2010; 118: 116-122.
- Matthews R, Azuero A, Asmellash S, Brewster E, Partridge EE, Piyathilake C. Usefulness of serum mass spectrometry to identify women diagnosed with higher grades of cervical intraepithelial neoplasia may differ by race. Int J Womens Health. 2011; 3: 185-192.