Research Article
A Proteomics. 2014;1(1): 6.
Characterization of Peptides and Low Molecular Weight Proteins in Plasma from Subjects with Hepatocellular Carcinoma
Xianyin Lai1,2, Frank A Witzmann2 and Suthat Liangpunsakul3,4*
1Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, USA
2Department of Cellular & Integrative Physiology, Indiana University School of Medicine, USA
3Department of Medicine, Indiana University School of Medicine, USA
4Roudebush Veterans Administration Medical Center, USA
*Corresponding author: Suthat Liangpunsakul, Department of Medicine, Division of Gastroenterology and Hepatology, Indiana University School of Medicine, 550 N, University Blvd, UH 4100, Indianapolis, IN 46202-5124, USA
Received: July 23, 2014; Accepted: August 25, 2014; Published: September 01, 2014
Abstract
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world with poor prognosis. Diagnosis of early stage of HCC is of importance to improve the prognosis and treatment outcome. The currently recommended biomarker alpha-fetoprotein (AFP), liver ultrasound (US),multi-detector row computed tomography (CT) scan, magnetic resonance imaging (MRI), and positron emission tomography (PET) have limitations in sensitivity, specificity, and tumors with a small size. Therefore, better plasma markers are needed for the early detection of HCC. The low molecular weight (LMW) fraction of plasma is made up of several classes of physiologically important proteins such as cytokines, chemokines, peptide hormones, as well as proteolytic fragments of larger proteins. To enrich the peptides and LMW proteins in plasma, 13 buffer systems were compared in the disruption of peptide/protein-protein interactions. A new two-step enrichment method using ultra filtration and solid-phase extraction (SPE) was developed and applied in the analysis of peptides and LMW proteins in hepatocellular carcinoma plasma samples. Statistical comparisons conducted within the PG600 software determined that 132 of 4,182 spectral peaks were found to differ between the control and HCC groups (p ≤ 0.05).
Keywords: Peptide; Low molecular weight protein; Plasma; Hepatocellular Carcinoma; MALDI-TOF
Introduction
Currently, hepatocellular carcinoma (HCC) is the fifth most common cancer in the world. The worldwide age-adjusted incidence of HCC in 2001 was shown to be 21 per 100,000 people and the age-adjusted mortality rate is 20.2 per 100,000 individuals [1,2]. The incidence and mortality rates for HCC are virtually identical, indicating the overall poor prognosis of this tumor. In the United States, HCC was the tumor with the largest increase in incidence over the past decades [1]. Liver cirrhosis is the most important factor in the development of HCC; therefore, patients with cirrhosis comprise the high risk group. HCC is an aggressive tumor, and median survival following diagnosis is approximately 6 to 20 months. Although the mainstay of therapy is surgical resection, the majority of patients are not eligible because of tumor extent due to late diagnosis or underlying liver dysfunction secondary to cirrhosis. Systemic chemotherapy is usually not well tolerated by patients with severe liver disease [1]. Therefore, diagnosis of early stage of HCC is of importance to improve the treatment outcome and prognosis.
Alpha-fetoprotein (AFP) is the only plasma marker currently recommended for HCC surveillance among patients with cirrhosis [1,2]. Prospective studies assessing AFP as a surveillance tool show a sensitivity of 39-65%, specificity of 76-91%, and a positive predictive value of 9-35% for early HCC. Liver ultrasound (US) has been reported to have a sensitivity of 80%, specificity of 91%, and a positive predictive value of 70% for the detection of early HCC in clinical studies [1]. However, the accuracy of US is operator-dependent, which limits its value as a surveillance test [1], and its use is also limited in obese patients. Though the sensitivity and specificity of multi-detector row CT scan and MR imaging with gadolinium are better than those with US, they appear to be sub-optimal diagnostic tools, especially for detection of tumor lesions < 2 cm. Other new imaging technology also failed to detect the early stages of hepatoma. In our recent study, we found that positron emission tomography (PET) has no role in detecting occult HCC in high-risk patients with hepatitis C awaiting liver transplantation [3]. This illustrates the need for better plasma markers for the detection of HCC.
Plasma is the most generally informative proteome from a medical view point to improve early disease detection [4], but it is a highly complex sample that contains proteins over a wide range of concentrations and molecular weights. The concentration of proteins in plasma varies from less than pg/mL to more than mg/mL [4]. Their molecular weight can be lower than 10 kDa or higher than 100 kDa. The most abundant proteins range from 50 kDa to 100 kDa [5], but the low molecular weight(LMW) fraction is made up of several classes of physiologically important proteins such as cytokines, chemokines, peptide hormones, as well as proteolytic fragments of larger proteins [6]. However, identification of peptides and LMW proteins is analytically challenging due to their low abundance in un depleted plasma or their absence in depleted plasma. Depletion of high abundance proteins concomitantly removes peptides and LMW proteins through non-covalent interactions with albumin and other carrier proteins in plasma [6, 7].
To enrich the peptides and LMW proteins in plasma, several approaches have been applied, such as gel chromatography and precipitation [8]. Villanueva et al. has reported a solid-phase extraction (SPE) method to capture peptides and LMW proteins [9,10]. Magnetic beads derivatized with reversed-phase ligands on the surface are mixed with the sample and washed with 0.1% TFA in water. Peptides and LMW proteins bound to the beads are eluted with 70% acetonitrile. The reversed-phase SPE method is efficient to capture and concentrate peptides and LMW proteins. However, it is polarity-specific and not size-specific. Many high molecular weight proteins can be captured by reversed-phase as well. Alternatively, centrifugal ultra filtration is more size-specific.
In centrifugal ultra filtration, peptide/protein-protein interactions have to be disrupted, so LMW components bound to larger species are released and free to pass through the molecular weight cut-off membrane [6,7]. This is due to the fact that carrier proteins in plasma bind physiologically important species such as hormones, cytokines, and lipoproteins [6]. Various buffer systems have been tested for the disruption of these interactions. However, all of the results indicate high molecular weight proteins appear in the filtrate. One of the reasons may be that the shape of some high molecular weight proteins changes after denature, leading their size change and making them pass through the membrane. Therefore, centrifugal ultra filtration and reversed-phase SPE should be combined to improve the enrichment of peptide and LMW proteins.
In this study to identify the unique markers in subjects with hepatocellular carcinoma, 13 buffer systems were compared in the disruption of peptide/protein-protein interactions. A new two-step enrichment method using ultra filtration and SPE was developed and applied in the analysis of peptides and LMW proteins in hepatocellular carcinomaplasma samples.
Materials and Methods
Ammonium bicarbonate (NH4HCO3), acetic acid (AA), formic acid (FA), trifluoroacetic acid (TFA), isopropyl alcohol (IPA), ethanol, methanol and urea were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN) and mass spectrometry-grade water were obtained from EMD Chemicals (Gibbstown, NJ, USA). Amicon Ultra-0.5 mL Centrifugal Filter (10K) was purchased from Millipore (Billerica, MA, USA).The a-cyano-4-hydroxycinnamic acid (CCA) and Oasis® HLB SPE plates were purchased from Waters Corporation (Milford, MA, USA).
Materials
Ammonium bicarbonate (NH4HCO3), acetic acid (AA), formic acid (FA), trifluoroacetic acid (TFA), isopropyl alcohol (IPA), ethanol, methanol and urea were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN) and mass spectrometry-grade water were obtained from EMD Chemicals (Gibbstown, NJ, USA). Amicon Ultra-0.5 mL Centrifugal Filter (10K) was purchased from Millipore (Billerica, MA, USA).The α-cyano-4-hydroxycinnamic acid (CCA) and Oasis® HLB SPE plates were purchased from Waters Corporation (Milford, MA, USA).
Study cohort
This study included 68 subjects (33 cirrhotic controls and 35 Cirrhotic subjects with hepatocellular carcinoma -HCC). All potential subjects were recruited from the pre-liver transplantation clinic at the Indiana University Hospital. Demographic, previous medical history and clinical information were obtained at the time of enrollment. The diagnosis of HCC was made by histopathology, and if histopathology is not available, the diagnosis was made by radiographic imaging (either magnetic resonance imaging [MRI], or dual-phase computed tomography [CT]) showing a vascular enhancing mass >1 cm and AFP > 200 ng/ml [as defined by the American Association for the Study of Liver diseases guideline[11]. In the cirrhotic controls without HCC, diagnosis of cirrhosis was based on liver histology or clinical, laboratory and imaging evidence of hepatic de compensation or portal hypertension [11]. To ensure that subjects with cirrhosis do not have occult HCC at the time of enrollment, these patients must have normal AFP (< 20 ng/ml) and normal CT/MRI. They were also prospectively followed for at least one year as a routine visit in the pre-transplantation clinic and repeated CT scan or MRI was done as part of HCC surveillance (according to the protocol for HCC screening/surveillance). At enrollment, 7 mL of blood were drawn and centrifuged at 1500 x g for 10 min at room temperature, within 1-3 hour(s) after acquisition. Aliquots (0.5 mL) were placed in separate 2 mL cryovials and stored at -80°C until analyses.
Centrifugal ultra filtration
The enrichment of peptides and LMW proteins in plasma was performed with a 10K cut-off centrifugal ultra filtration filter. The centrifugal filter was used according to the manufacturer's specifications. Thirty six micro liters of plasma were diluted with 180 μLof 20% (v/v) acetic acid. The diluted sample (216 μL) was added to an Amicon Ultra-0.5 mL Centrifugal Filter (MWCO 10K), and centrifuged at 3,000 x g for 40 min. After the first centrifugation, 180 μL of 20% (v/v) acetic acid were added to the A micron Ultra filter device. The sample was centrifuged at 3,000 x g for 40 min again. The remaining is about 50 μL. The filtrate was dried with a Speed Vacuum.
Reversed-phase SPE
The purification of extracted peptides and protein was conducted with an OASIS Zip Plate and vacuum. The dried sample was reconstituted with 50 μL of water with 0.1% FA (v/v), loaded on an equilibrated Zip Plate, and incubated for 8 min. The Zip Plate was washed three times with 100 μL of 5% acetonitrile in water with 0.1% FA. The sample was eluted from the Zip Plate with 50 μL of 70% acetonitrile in water with 0.1% FA twice. The eluate was dried with a Speed Vacuum for 2 hands the sample was reconstituted with 20 μL of 50% acetonitrile in water with 0.5% FA.
MALDI-TOF Mass Spectrometry
Mass spectra were acquired on a Bruker Auto flex III MALDI-TOF mass spectrometer interfaced with Bio Tools™ software suite from Bruker Daltonics. For calibration of the instrument, 1 μL of the 4700 mix solution was mixed with 5 μL of 10 g/L solution of a-cyano-4-hydroxycinnamic acid (CCA) in 10:9:1 water: acetonitrile: acetone containing 0.1% trifluoroacetic acid. For this study, 1 μL of the sample was combined with 1 μL of a 10 g/L CCA solution. 1 μL of the mixed solution was deposited onto the target plate and allowed to dry. The target plate was introduced into the mass spectrometer and irradiated with a 337 nm laser at intensity sufficient to just ionize the sample. Mass spectra were recorded from700 to 4,000 Daunder reflector modes and from 4,000 to 12,000 Da under linear mode. Each sample had two mass spectra. Typical resolution was 11,000 at 3.2 kDa under reflector mode and 4,000 at 3.2 kDa (1,000 at12 kDa) under linear mode.
Processing and analysis of MALDI spectra
MALDI mass spectral data were processed and analyzed using Pro genesis PG600™ software (Non Linear Dynamics) essentially as described by Lopez et al. [12]. Raw spectra were loaded directly into the PG600 program using the prOTOF loader program. The individual MALDI peak quantity was determined as the integral of the corrected intensity over the width of the peak envelope. The raw signal was filtered to remove noise and background signals by a discrete wavelet transform. Peaks were then detected and the relationship between m/z value and the size (width and intensity) of the isotopic peak distribution determined by detecting the start and end of the peak envelope. The intensity of this envelope was then integrated over this range to give the Peak Quantity. The data were normalized by summing all spectral peaks, with the assumption that all spectra have approximately the same material in them. All sample spectra were processed in this manner, and analyses were performed on the 33 control and 35 hepatocellular carcinoma samples to find discriminate markers between the two groups. Parameters for biomarker selection were set within the PG600 program to include peaks with a mean quantity threshold of ≥ 100 and statistical confidence interval for treatment group differences set at p ≤ 0.05.
Results
Clinical characteristics of the study cohort
The characteristics and etiologies of underlying liver diseases of the study participants are presented in Table 1. In both cohorts, there were no differences in age and gender between groups. However, 90% of controls were Caucasian compared to 63% in those with HCC (p = 0.01).
Demographic and clinical characteristics
Study cohort
Controls
(n = 33 )
Cases with HCC (n = 35)
p-value
Age (yrs)
59.4±6.9
58±7.9
0.20
Male sex, %
69%
68%
0.12
Race, White (%)
90%
63%
0.01
Etiologies of liver diseases (N)
:Hepatitis C and alcohol
4
8
0.01
:Alcoholic liver disease
10
4
:Hepatitis C
8
13
:Non-alcoholic steatohepatitis
8
5
:Hepatitis B
-
5
Autoimmune hepatitis
3
-
Table 1: Clinical characteristics of the study cohort.
Peptides and low molecular weight proteins enrichment
In plasma, twenty-two high abundance proteins constitute about 99% of the protein content [6]. Peptides and LMW proteins are low-abundant in plasma. Therefore, it is difficult to identify them in un depleted plasma. Affinity depletion is a popular method to remove highly-abundant proteins, but one of the fundamental drawbacks of this method is that many important peptides and LMW proteins might be concomitantly removed by the sample preparation process [13]. Hence, a solution to disrupt protein-protein interactions should be applied to release LMW components bound to carrier proteins before the depletion of high abundant proteins. In this situation, only the centrifugal ultra filtration method conducted under dissociating solutions is the ideal depletion method to remove highly abundant proteins but leave remaining those peptides and proteins bound to them. Accordingly, the dissociating solution is the most important factor that affects the recovery of LMW species.
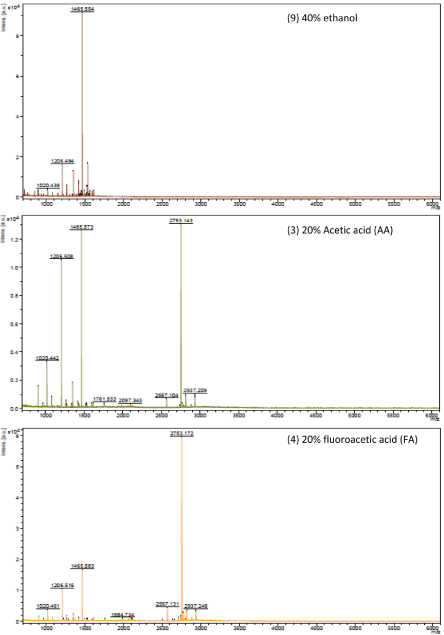
To assess the effect of disrupting of protein-protein interactions, 13 dissociating solutions were applied in this study: (1) 25mM NH4HCO3, pH 8.2, 20% (v/v) acetonitrile, (2) 4 M urea + 25mM NH4HCO3, pH 8.2, 20% (v/v) acetonitrile, (3) 20% Acetic acid (AA), (4)20% fluoroacetic acid (FA), (5) 10% trifluoroacetic acid (TFA), (6) 20% methanol, (7) 40% methanol, (8) 20% ethanol, (9) 40% ethanol, (10) 20% isopropyl alcohol (IPA), (11) 40% isopropyl alcohol (IPA), (12) 10% Acetonitrile (ACN), (13) 20% Acetonitrile (ACN). Solutions (1) and (2) are from previous publications [6,7]. The spectra of samples processed with the 13 dissociating solutions were compared. Three representative spectra are illustrated in Figure 1. Two main peak clusters are shown in the spectra. One includes peaks around 1465 Da (peak cluster 1) and the other one includes peaks around 2753 Da (peak cluster 2). They form three typical patterns: 1) peak cluster 1 is highly-abundant while peak cluster 2 is extremely low-abundant, 2) both peak clusters 1 and 2 are highly-abundant, and 3) peak cluster 1 is low-abundant while peak cluster 2 is highly-abundant. Among the 13 dissociating solutions, #1-2, 5, and 6-13 generated samples of the pattern 1, #3 generated samples of the pattern 2, and #4 generated samples of the pattern 3. Acetic acid and fluoroacetic acid are more likely to break protein-peptide/protein interactions. According to the number and intensity of peaks, 20% acetic acid (AA) generated the best recovery of peptides and LMW proteins. Hence, AA is applied in the analysis of the samples.
Figure 1 : Three representative spectra of samples processed with the dissociating solutions (9) 40% ethanol, (3) 20% Acetic acid (AA), and (4) 20% fluoroacetic acid (FA). According to the number and intensity of peaks, 20% acetic acid (AA) generated the best recovery of peptides and LMW proteins.
As mentioned earlier, the centrifugal ultra filtration alone is not an ideal method because high molecular proteins pass through the membrane. To further enrich the peptides and LMW proteins after the centrifuge, the reverse-phase SPE method is applied. Therefore, a new two-step enrichment method was developed and applied in this study.
MALDI profiles of biomarker enriched samples
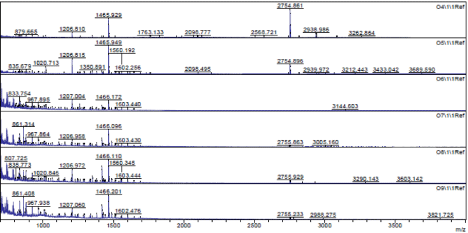
Potential biomarkers of peptides and low molecular weight proteins were enriched using the developed method and resulted in the detection of 4182 spectral peaks or features (from 700.10-11,833.91 Da) that were matched and compared between controls and those with hepatocellular carcinoma across all 68 samples. Typical MALDI profiles are illustrated in Figure 2. Statistical comparisons conducted within the PG600 software determined that 132 of these were found to differ (p ≤ 0.05); 117 features increase and 15 features decrease in subjects with hepatocellular carcinoma compared to the controls (see Figure 3 & Table 2).
Figure 2 : Matrix-assisted laser-desorption ionization time-of-flight (MALDITOF) mass spectrum of typical enriched samples.
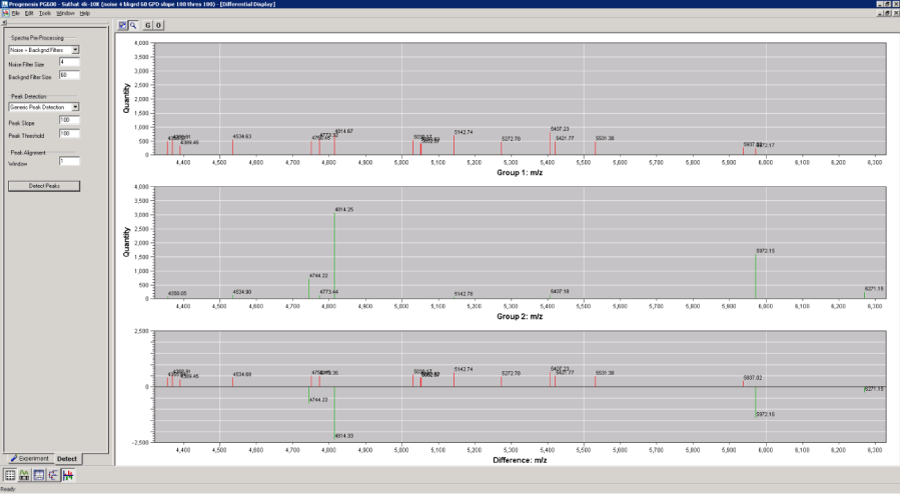
Figure 3 : PG600™ software comparison of spectral peaks from control and hepatocellular carcinoma samples that were detected, normalized, processed, and statistically compared. Upper panel shows mean control peak intensities, middle panel shows mean hepatocellular carcinoma, and the bottom panel shows the comparative difference of the significantly different biomarker candidates. X-axesare mass/charge (m/z) and Y-axes are normalized peak intensity calculated from ion current.
m/z
Control Quantity
HCC Quantity
Fold
P
Value
m/z
Control Quantity
HCC Quantity
Fold
p Value
701.52
5931.4
9965.7
-1.7
0.01
790.68
1072.6
1957.5
-1.8
0.01
702.52
2248.1
3824.3
-1.7
0.009
793.71
662.1
1142.4
-1.7
0.03
704.73
503.8
1255.6
-2.5
0.03
795.73
1480.3
3096.9
-2.1
0.01
705.53
1058.9
1777.3
-1.7
0.03
796.74
800.7
1524.4
-1.9
0.03
707.56
2307.3
5191.6
-2.3
0.005
797.70
437.4
793.5
-1.8
0.04
708.56
897.4
1878.3
-2.1
0.01
798.47
115.2
30.0
3.8
0.04
710.48
645.7
1752.7
-2.7
0.005
798.73
226.3
530.4
-2.3
0.03
711.42
1730.1
2621.7
-1.5
0.04
801.64
495.0
964.0
-2.0
0.04
713.44
2502.7
3928.8
-1.6
0.02
809.71
513.2
975.8
-1.9
0.03
714.56
207.1
1002.8
-4.8
0.02
811.70
535.0
1185.1
-2.2
0.01
716.57
531.7
1404.3
-2.6
0.01
813.72
118.4
475.5
-4.0
0.01
718.58
565.4
1250.2
-2.2
0.03
815.74
281.8
595.5
-2.1
0.04
719.53
1546.8
2548.3
-1.7
0.03
820.79
137.0
538.4
-3.9
0.009
721.53
1095.2
1955.8
-1.8
0.01
823.83
152.1
905.2
-6.0
0.002
722.59
743.6
1553.2
-2.1
0.01
829.68
1113.0
2032.0
-1.8
0.05
723.55
1423.8
2570.9
-1.8
0.03
830.76
121.8
590.9
-4.9
0.008
728.61
450.7
1277.4
-2.8
0.008
833.82
629.4
2157.0
-3.4
0.003
729.52
731.2
1789.8
-2.5
0.01
834.73
755.6
1406.2
-1.9
0.01
730.61
158.3
962.0
-6.1
0.003
839.76
1052.4
2010.8
-1.9
0.03
733.57
1118.5
2018.7
-1.8
0.04
847.76
241.2
747.4
-3.1
0.01
735.65
1311.8
2675.4
-2.0
0.01
848.76
538.9
956.8
-1.8
0.02
736.43
57.9
0.0
Infinite
0.02
851.74
906.4
1514.2
-1.7
0.04
736.66
972.0
1765.8
-1.8
0.01
857.79
75.0
224.9
-3.0
0.04
737.70
859.0
1801.8
-2.1
0.05
861.44
202.3
2170.3
-10.7
0.03
739.67
214.5
1722.4
-8.0
0.0005
864.79
507.8
873.6
-1.7
0.05
740.55
1053.5
1827.6
-1.7
0.02
873.71
611.6
1243.6
-2.0
0.03
741.67
876.2
3280.3
-3.7
0.02
875.83
115.5
470.1
-4.1
0.02
742.61
1427.4
2609.0
-1.8
0.01
877.75
893.5
1693.4
-1.9
0.01
743.63
1318.7
2168.8
-1.6
0.03
883.81
834.1
1677.6
-2.0
0.008
745.63
3720.6
6454.8
-1.7
0.02
887.79
54.8
201.3
-3.7
0.04
746.64
1634.2
2824.0
-1.7
0.01
894.13
154.6
27.8
5.6
0.05
748.80
424.2
1245.2
-2.9
0.05
894.86
25.1
143.7
-5.7
0.04
750.72
735.1
1279.3
-1.7
0.03
895.28
65.6
0.0
Infinite
0.02
751.73
1628.1
4084.8
-2.5
0.02
916.54
24.2
0.0
Infinite
0.05
752.48
51.7
0.0
Infinite
0.05
918.87
69.3
251.3
-3.6
0.04
752.69
1088.7
2222.2
-2.0
0.005
921.79
566.8
1120.1
-2.0
0.03
753.65
688.2
1246.2
-1.8
0.02
963.88
127.0
383.4
-3.0
0.04
755.54
842.8
1509.4
-1.8
0.02
1006.66
39.2
0.0
Infinite
0.03
756.63
431.8
993.0
-2.3
0.02
1009.91
567.5
1250.1
-2.2
0.02
757.54
1451.4
2336.2
-1.6
0.02
1031.81
56.5
0.0
Infinite
0.02
759.64
966.5
1671.9
-1.7
0.03
1054.03
0.0
97.6
Infinite
0.03
762.61
695.3
1459.8
-2.1
0.004
1206.97
1787.2
4584.4
-2.6
0.04
763.62
958.9
1719.4
-1.8
0.05
1282.76
82.2
8.6
9.5
0.05
764.67
600.1
1221.7
-2.0
0.03
1420.04
997.9
2836.8
-2.8
0.04
765.67
711.1
1422.2
-2.0
0.02
1466.03
18456.9
30130.2
-1.6
0.05
767.69
877.1
1876.1
-2.1
0.01
1488.03
1190.5
2261.0
-1.9
0.05
768.67
705.3
1260.8
-1.8
0.03
1511.26
0.0
69.2
Infinite
0.03
769.76
66.3
501.0
-7.6
0.01
4355.64
478.9
69.9
6.9
0.05
771.66
783.5
1353.6
-1.7
0.02
4368.91
526.5
0.0
Infinite
0.04
772.57
234.5
599.2
-2.6
0.04
4389.45
331.5
0.0
Infinite
0.05
773.66
448.3
944.7
-2.1
0.02
4534.68
540.2
123.6
4.4
0.04
774.65
212.8
880.8
-4.1
0.002
4744.22
0.0
714.7
Infinite
0.04
775.69
996.4
1722.7
-1.7
0.05
4750.45
491.6
0.0
Infinite
0.04
776.77
295.5
832.6
-2.8
0.02
4773.35
589.2
123.1
4.8
0.05
777.70
652.0
1201.5
-1.8
0.03
4814.33
720.7
3072.3
-4.3
0.05
778.72
402.1
880.3
-2.2
0.04
5030.17
527.4
0.0
Infinite
0.04
779.45
85.4
10.3
8.3
0.03
5050.82
439.0
0.0
Infinite
0.02
779.69
977.9
1885.2
-1.9
0.008
5052.87
384.0
0.0
Infinite
0.05
780.69
744.5
1270.2
-1.7
0.03
5142.74
697.2
60.4
11.5
0.04
782.52
66.1
272.2
-4.1
0.02
5272.78
452.8
0.0
Infinite
0.04
783.62
439.3
160.3
2.7
0.03
5407.23
811.1
149.5
5.4
0.05
783.73
352.8
1244.0
-3.5
0.004
5421.77
485.2
0.0
Infinite
0.02
785.71
551.6
2335.5
-4.2
0.006
5531.38
481.7
0.0
Infinite
0.05
787.66
867.0
1563.8
-1.8
0.03
5937.02
263.5
0.0
Infinite
0.05
788.66
478.2
933.6
-2.0
0.02
5972.16
224.2
1604.2
-7.2
0.02
789.67
2398.1
4213.2
-1.8
0.02
6271.15
0.0
241.2
Infinite
0.05
Table 2: Potential biomarkers detected by MALDI mass spectrometry in control and HCC samples.
Discussion
The objective of this project was to detect novel bio markers of hepatocellular carcinoma (HCC). We hypothesized that HCC elicits cellular and molecular responses whose sequelae are revealed through the appearance of unique and low-abundance polypeptides or proteins in the plasma. As a result of the comprehensive mass spectrometry-based analysis used in this pilot study, we have identified biomarker candidates whose presence in plasma is altered in HCC subjects compared to controls. Identification and relative quantitation of some of these peptides and low molecular weight proteins with respect to clinical data supports our hypothesis that the peptides and low molecular weight proteins can be used to detect the presence of liver cancer.
It is important to identify the 132 peaks that differed between the control and HCC groups. Due to the limitation of our instrument, MALDI-TOF/TOF data was not available. Follow-up work should focus on the identification of the 132 peaks and their statistical analysis to discover biomarker candidates with good specificity and sensitivity.
In summary, we have shown in this study that potential protein markers have appeared in mass spectral profiles and that they may be useful clinically to determine hepatocellular carcinoma by MALDI-TOF mass spectrometry. However, a large-scale study is needed to confirm and validate our current results.
Acknowledgement
This work was supported by a grant from IU Health/Clarian Value fund (VFR 260), K08 AA016570 from the NIH/NIAAA, 1I01CX000361-01 from the Veterans Affairs Research and Administration.
References
- Marrero JA. Screening tests for hepatocellular carcinoma. Clin Liver Dis. 2005; 9: 235-25.
- Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, Comunale MA. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005; 43: 1007-1012.
- Liangpunsakul S, Agarwal D, Horlander JC, Kieff B, Chalasani N. Positron emission tomography for detecting occult hepatocellular carcinoma in hepatitis C cirrhotics awaiting for liver transplantation. Transplantation proceedings. 2003; 35: 2995-2997.
- Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004; 3: 311-326.
- Bellei E, Bergamini S, Monari E, Fantoni LI, Cuoghi A, Ozben T, et al. High-abundance proteins depletion for serum proteomic analysis: concomitant removal of non-targeted proteins. Amino Acids. 2011; 40: 145-156.
- Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003; 2: 1096-1103.
- Jung WW, Phark S, Oh S, Khim JY, Lee J, Nam MH, et al. Analysis of low molecular weight plasma proteins using ultrafiltration and large gel two-dimensional electrophoresis. Proteomics. 2009; 9: 1827-1840.
- Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006; 6: 6326-6353.
- Villanueva J, Philip J, Entenberg D, Chaparro CA, Tanwar MK, Holland EC, et al. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal Chem. 2004; 76: 1560-1570.
- Villanueva J, Lawlor K, Toledo-Crow R, Tempst P. Automated serum peptide profiling. Nat Protoc. 2006; 1: 880-891.
- Bruix J, Sherman M. Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology. 2005; 42: 1208-1236.
- Lopez MF, Mikulskis A, Kuzdzal S, Golenko E, Petricoin EF, Liotta LA, et al. A novel, high-throughput workflow for discovery and identification of serum carrier protein-bound peptide biomarker candidates in ovarian cancer samples. Clin Chem. 2007; 53: 1067-1074.
- Huang HL, Stasyk T, Morandell S, Mogg M, Schreiber M, Feuerstein I, et al. Enrichment of low-abundant serum proteins by albumin/immunoglobulin G immunoaffinity depletion under partly denaturing conditions. Electrophoresis. 2005; 26: 2843-2849.