
Research Article
Austin J Pulm Respir Med. 2023; 10(2): 1100.
A Correlative Study on EGFR Gene Mutation Status of Primary Lung Tumor and Brain Metastases in NSCLC
Jinrong Hu¹*; Jinlian Hu²
¹West China School of Public Health, Sichuan University/Department of Palliative Medicine, West China Fourth Hospital, Chengdu 610000, P.R. China
²Rheumatology and Immunology Department of Deyang People’s Hospital, Deyang 618000 PR China
*Corresponding author: Jinrong Hu West China School of Public Health, Sichuan University/Department of Palliative Medicine, West China Fourth Hospital, Chengdu 610000, P.R. China. Tel: 15928943007 Email: 710459182@qq.com
Received: October 21, 2023 Accepted: November 29, 2023 Published: December 06, 2023
Abstract
Objective: This study aims to explore the predictive role of EGFR gene mutation in the primary lung tumor on brain metastases in Non-Small Cell Lung Cancer (NSCLC). Methods: Morphological differences between EGFR gene 19 exon deletion mutant HCC827 and wild-type EGFR gene A549 human lung adenocarcinoma cells were observed under a microscope. The MTT method was employed to detect the proliferation differences between HCC827 and A549 cells. The Transwell in vitro cell invasion experiment was used to compare the invasion capabilities of the two cell lines. Furthermore, the Χ2 test was applied to analyze the relationship between the EGFR mutation in the primary lung tumor and the occurrence of brain metastases in 253 NSCLC patients. Results: A549 cells were smaller in size, resembling paving stones, whereas HCC827 cells were larger and polygonal. MTT analysis revealed that wild-type EGFR A549 human lung adenocarcinoma cells proliferated faster than EGFR mutant HCC827 cells, with a significant difference (P<0.05). The Transwell in vitro cell invasion experiment indicated that HCC827 cells were significantly stronger than A549 cells (P<0.05). The incidence of brain metastases in 253 NSCLC patients was 12.3% (31/253), among which eight cases had EGFR gene mutations in primary lung tumor, with a mutation rate of only 25.81% (8/31). No significant correlation was found between the occurrence of brain metastases and the mutation in primary lung tumor. Conclusion: EGFR gene mutation in NSCLC cells can significantly enhance their invasive activity. However, the correlation between EGFR gene mutation in primary lung tumor and the occurrence of brain metastases warrants further study.
Keywords: Non-small cell lung cancer; EGFR gene; Epidermal Growth Factor Receptor; Brain Metastases
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with Non-Small Cell Lung Cancer (NSCLC) accounting for approximately 80% of all lung cancers. The incidence of brain metastases in NSCLC disease progression is as high as 25% to 54%, often presenting as multiple metastases [1]. ATP competitive tyrosine kinase inhibitors (Tyrosine Kinase Inhibitors, TKIs) such as Erlotinib, Gefitinib, and Icotinib, which target the Epidermal Growth Factor Receptor (EGFR), are now widely used in the clinical treatment of NSCLC. It is widely agreed that EGFR-activating mutations are the best predictors of the efficacy of EGFR-TKIs in NSCLC patients [2-4]. However, the acquisition of satisfactory tumor tissue samples for EGFR mutation detection is often hindered by numerous objective factors. It is well known that EGFR mutant lung cancer is a special type of NSCLC, where the cancer cells of this type of lung cancer rely on the "EGFR pathway" to maintain growth, proliferation, and metastasis. Studies have shown that brain metastases in NSCLC may be related to its EGFR mutation[5]. Therefore, this project intends to explore the predictive effect of NSCLC brain metastases on primary lung tumor EGFR mutations through clinical samples and in vitro experiments, aiming to provide a theoretical basis for screening populations that are effective for TKI treatment.
Materials and Methods
Clinical data
This study collected 253 cases of newly diagnosed NSCLC patients from the Daping Hospital, Military Medical University from July 2011 to May 2013. The inclusion criteria were histologically confirmed NSCLC, patients completing head MRI examination within 2 weeks before and after obtaining specimens, lung cancer clinical staging performed according to the 2009 seventh edition TNM staging criteria, the paraffin-embedded specimens from the primary lung tumor met the requirements of the ARMS method for EGFR mutation detection, and informed consent was signed with each patient/relative. Exclusion criteria included patients with a second malignant tumor, patients with severe pulmonary fibrosis, patients who had previously received radiation therapy or chemotherapy, and pregnant or nursing women.
Experimental materials
Modified RPMI-1640 culture (Hyclone), Australian fetal bovine serum (Gibco), trypsin (Gibco), MTT (Gibco), PBS (Meixin), Cell Cryoprotectant DMSO (sigma), DMSO (amresco), 50ml culture flask (corning), 48-well plate (biofil), 24-well plate (biofil), transwell 24-well 8μm (corning), 15ml centrifuge tube (kirgen), A549 cells (External Research Institute Room 1), HCC827 cells (gifted by Professor He Yong, Department of Respiratory Medicine, Daping Hospital). Instrument provided by Field Surgery Research Institute of Daping Hospital, Military Medical University were: ELISA detector (BIOTEK, ELX800), cell culture box, inverted microscope.
EGFR gene mutation analysis
The Amplification Refractory Mutation System (ARMS) was used to detect EGFR-TK domain mutations. The Human EGFR Gene Mutation Fluorescence PCR Assay Kit was used according to the ADx-ARMS EGFR Mutation Test Kit (AmoyDx) protocol, which can detect 29 types of EGFR gene mutations including T790M, G719S, G719A, G719C, L858R, L861Q, S7681, 3 types of exon 20 insert mutations, and 19 types of exon 19 deletion mutations.
Cell culture
A549 and HCC827 cells were cultured in RPMI1640 supplemented with 10% fetal bovine serum, 100μ/L penicillin and 100μg/L streptomycin, in a 37o 5%CO2 cell incubator, and the cell morphology was observed.
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) method to detect cell proliferation
0.5g MTT was dissolved in 100ml PBS, filtered through a 0.22um filter, and stored at -20°C. Under an inverted microscope, when both types of cells were in the logarithmic growth phase, the culture medium was removed and serum-free medium was added for starvation for 12 hours. After cell cycle synchronization, cells were digested with trypsin and collected. After cell counting, 1x10^4 cells were added to each well. Six secondary wells were set up for each group of cells, and 50μL MTT was added at 12h, 48h, 72h and 96h. After 4 hours, the culture medium was removed and 300μL DMSO was added to dissolve the formazan. After 15 minutes, After 15 minutes, the absorbance was measured at a wavelength of 490nm (OD490). The same experiment was repeated three times.
Transwell cell invasion assay in vitro
1) The cell culture flask was taken out of the CO2 incubator and observed under an inverted microscope. When the cells were in the logarithmic growth phase, the culture medium in the flask was replaced with serum-free medium. After starving the cells for 12 hours, the cells were digested with trypsin, and after digestion, the cells were centrifuged and the culture medium was discarded (washed once with PBS), and then resuspended in culture medium containing 5% FBS. The cell density was adjusted to 5x10^5 cells/ml. 2) After the matrix gel was thawed on ice, double the volume of serum-free culture medium was added, and then mixed with a 200ul pipette tip. 50ul of the mixture was added into each Transwell pore. After 30 minutes at 37°C, the matrix gel was polymerized into a gel and set aside. 3) 200ul of cell suspension was added to the small chamber, and 600ul of 20% FBS culture medium was added to the lower chamber of the 24-well plate. The plate was then incubated in a cell culture incubator at 37°C and 5% CO2 for 48 hours. 4) After incubation, the Transwell chamber was removed, the culture medium in the pore was discarded, and the chamber was washed twice with calcium-free PBS. The unmigrated cells on the upper layer were gently wiped off with a cotton swab, and the chamber was inverted to air dry. The Transwell chamber was placed into a clean 24-well plate, and 600 μl of 0.1% crystal violet solution diluted with methanol was added to each well to submerge the membrane in crystal violet. After 30 minutes at room temperature, the chamber was removed and washed three times with PBS. Under a 200x microscope, six random fields of view were selected to observe the cells, photos were taken, and the cells were counted. The average was calculated, and each group had three replicate wells. The mean and standard deviation were calculated. The same experiment was repeated three times.
Statistical analysis
Experimental data are expressed as mean ± standard deviation (x±s), analyzed using SPSS17.0 statistical software, the a value for hypothesis testing is 0.05, P<0.05 indicates statistically significant differences. The comparison of MTT OD values, the number of migrated cells, and the number of invaded cells between the two different cell types was analyzed using one-way ANOVA.
Results
Clinical features of NSCLC patients with brain metastases and EGFR gene mutation in primary lung tumor
Out of 253 NSCLC patients, 31 had brain metastases, with a metastasis rate of 12.3% (31/253). Among the 31 NSCLC patients with brain metastases, only 8 had EGFR gene mutations in primary lung tumor, with a mutation rate of 25.81% (8/31). While in the 222 NSCLC patients without brain metastases, 99 had EGFR gene mutations, with a mutation rate of 44.59% (99/222). There was a significant difference between the two groups (P<0.05), as shown in Table 1, with more EGFR mutations in primary lung tumor of NSCLC patients without brain metastases.
Patients with brain metastases
Patients without brain metastases
N
%
N
%
EGFR mutation
8
25.81%
99
44.59%
EGFR wild-type
23
74.19%
123
55.41%
P
0.047
Table 1: Relationship between EGFR mutation in primary lung tumor and brain metastases.
Comparison of morphologies of EGFR wild-type and mutated lung cancer cells
When both types of cells grow to the logarithmic phase, observation under an inverted microscope at a magnification of 100x shows that the EGFR mutated HCC827 cells are larger and polygonal (Figure 1A); while the wild-type EGFR A549 cells are smaller and resemble paving stones (Figure 1B).
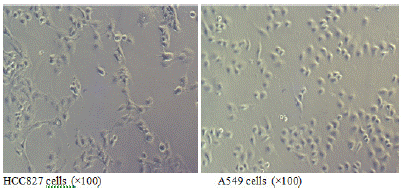
Figure 1: Morphology of EGFR mutated HCC827 cells and EGFR wild-type A549 cells.
EGFR wild-type lung cancer cells proliferate faster than EGFR mutated lung cancer cells
After starving EGFR mutated HCC827 cells and wild-type EGFR A549 cells for 12 hours, and after cell cycle synchronization, they were seeded in equal numbers into a 48-well plate. It can be seen that the proliferation rate of the EGFR wild-type A549 cells is significantly faster than that of the EGFR mutated HCC827 cells (Table 2, Figure 2).
Groups
12H
48H
72H
96H
HCC827
0.159±0.014
0.284±0.011
0.379±0.017
0.528±0.023
A549
0.149±0.008
0.311±0.013
0.501±0.37
0.669±0.018
P
0.171
0.003
<0.001
<0.001
Table 2: Comparison of proliferation activity between EGFR mutated HCC827 cells and EGFR wild-type A549 cells.
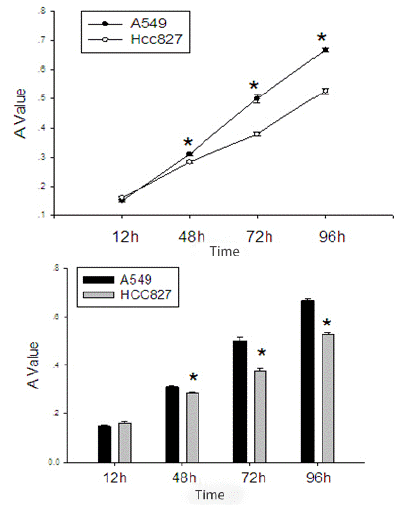
Figure 2: Comparison of proliferation activity between EGFR mutated HCC827 cells and EGFR wild-type A549 cells.
*Indicates P value<0.05 compared with EGFR mutated HCC827 cells and EGFR wild-type A549 cells
EGFR mutated lung cancer cells have stronger invasion ability than EGFR wild-type lung cancer cells
After seeding an equal number of EGFR mutated HCC827 cells and wild-type EGFR A549 cells onto the upper chamber of a Transwell coated with matrix gel, an observation after 48 hours shows that the ability of HCC827 to invade through the matrix gel is significantly stronger than A549 (Table 3, Figure 3).
Groups
Group 1
Group 2
Group 3
HCC827
227±37
199±14
207±10
A549
39±5
48±8
51±4
P
0.001
<0.001
<0.001
Table 3: Comparison of invasion ability between EGFR mutated HCC827 cells and EGFR wild-type A549 cells.
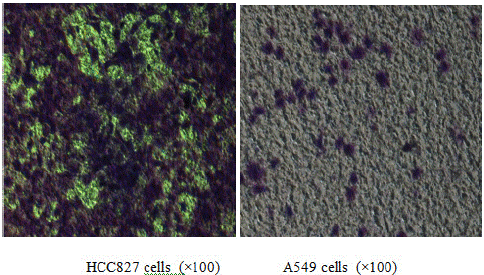
Figure 3: Comparison of invasion activity between EGFR mutated HCC827 cells and EGFR wild-type A549 cells.
Discussion
The activation of the EGFR signaling pathway can be broadly divided into: 1) an increase in receptor expression; 2) continuous activation due to receptor mutations that are not ligand-dependent; 3) through autocrine ligand secretion. High expression of EGFR is common in advanced NSCLC, and 65% of lung adenocarcinoma cells overexpress EGFR. Upon activation, the EGFR signaling pathway typically promotes cell proliferation and metastasis, whilst inhibiting apoptosis of tumor cells via signal transduction pathways such as K-ras, PIK3, and STAT3/5 [6]. Morphologically, cells with EGFR gene mutations are larger and more angular, which is more conducive for tumor cells to extend pseudopodia to invade organs and pass through the basement membrane in an amoeboid manner compared to smaller, cobblestone-like wild-type EGFR lung adenocarcinoma cells [7]. Our experiments also confirmed that lung adenocarcinoma cells with EGFR mutations have significantly stronger invasive abilities than wild-type EGFR cells. This aligns with literature reports [8] that high EGFR expression in primary NSCLC lesions is closely related to tumor invasiveness. In terms of proliferation, A549 cells have significantly stronger proliferative abilities than EGFR-mutated cells HCC827, which may be related to the K-ras gene mutation in A549 lung adenocarcinoma cells. We know that K-ras gene mutations can cause continuous activation of the EGFR signaling pathway, leading to the continuous growth of malignant tumors, and K-ras gene mutations can make NSCLC with EGFR gene mutations insensitive to EGFR inhibitors.
Relevant literature reports [9] that among patients who have had stage I lung adenocarcinoma removed, tumor diameter and EGFR gene mutation are independent factors affecting postoperative recurrence or metastasis in lung adenocarcinoma patients. Patients with tumors larger than 2cm or with wild-type EGFR genes have a significantly higher risk of recurrence than patients with tumors smaller than 2cm or EGFR-mutated genes. However, in late-stage NSCLC, EGFR gene mutations are commonly found in tissues from brain metastases and primary lung tumor [10]. From our experiments, wild-type EGFR lung adenocarcinoma cells proliferate significantly faster than EGFR-mutated cells, but the invasive ability of cells is significantly stronger in EGFR-mutated types than in wild types. This may be related to the stronger proliferative ability of early-stage lung adenocarcinoma cells with wild-type EGFR genes, and the local recurrence of early-stage tumors is closely related to their proliferative ability, while late-stage lung cancer cells with EGFR gene mutations increase their invasive ability and enhance their ability to metastasize to distant places.
There have been a few reports [10,11] on the correlation between the occurrence of brain metastases in Non-Small Cell Lung Cancer (NSCLC) and EGFR gene mutations in tumor cells at the primary site, but the mechanism of their occurrence has not yet been clarified. From our experimental results, we can see that patients with wild-type EGFR genes are more likely to have brain metastases. This is inconsistent with the results reported by Li [5] et al.They found that among 110 NSCLC patients, 14 had brain metastases, of which 9 had EGFR gene mutations in the primary lesions, while only 31.2% of the 96 patients without brain metastases had EGFR gene mutations in the primary lesions. The reasons for these differences are analyzed: 1) it may be related to different EGFR gene detection methods. Our experiment detects EGFR-TK structural domain mutations through the ARMs method, while they use sequencing to detect EGFR gene mutations. The detection sensitivity of the ARMs method is much higher than that of sequencing (1-2% vs. 10-20%) [9]. 2) The sample size is small. Li et al. reported that only 14 of 110 patients (13%) had brain metastases, while in our study, the probability of NSCLC brain metastases was 12.3%. The incidence of brain metastases in NSCLC reported in both studies is lower than the generally observed probability of 25%~54%. A meta-analysis of EGFR genes showed [12] that the number of copies of EGFR genes in metastatic lesions is higher than that in primary lesions, and there is no significant difference in the frequency of EGFR gene mutations between primary and metastatic lesions. Therefore, the relationship between the EGFR gene status and brain metastases may require a larger sample or detection methods with higher sensitivity and accuracy.
Author Contributions: Hu Jinrong, Hu Jinlian. Hu Jinrong. writing—original draft preparation and review and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement: This study was conducted in accordance with the Declaration of Helsinki and approved by the Medical Ethics Committee of Daping Hospital, Military Medical University (18.09.2013)
Informed Consent Statement: All patients gave their written informed consent to participate in this study.
Funding: This research received no external funding.
Data Availability Statement: Data supporting the findings of this investigation can be obtained from the corresponding author via appropriate request.
Acknowledgments: The authors wish to thank other members of our research group who work behind the scenes.
Conflicts of Interest: The authors declare no conflict of interest.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2008; 58: 71-96.
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004; 304: 1497-500.
- Sholl LM, Xiao Y, Joshi V, Yeap BY, Cioffredi LA, Jackman DM, et al. EGFR mutation is a better predictor of response to tyrosine kinase inhibitors in non–small cell lung carcinoma than FISH, CISH, and immunehistochemi-stry. Am J Clin Pathol. 2010; 133: 922-34.
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009; 361: 947-57.
- Li Z, Lu J, Zhao Y, Guo H. The restrospective analysis of the frequency of EGFR mutations and the efficacy of gefitinib in NSCLC patients with brain metastasis. J Clin Oncol. 2011; 29: e18065.
- Ganti AK. Epidermal growth factor receptor signaling in nonsmall cell lung cancer. Cancer Invest. 2010; 28: 515-25.
- Gao J. Experimental study of invasion and metastasis of cancer cells and establishment of the morphological pattern in the process of invasion and metastatic models of cancer cells. Sci Sin B. 1988; 31: 719-25.
- Izar B, Sequist L, Lee M, Muzikansky A, Heist R, Iafrate J, et al. The impact of EGFR mutation status on outcomes in patients with resected Stage I non-small cell lung cancers. Ann Thorac Surg. 2013; 96: 962-8.
- Burel-Vandenbos F, Ambrosetti D, Coutts M, Pedeutour F. EGFR mutation status in brain metastases of non-small cell lung carcinoma. J Neurooncol. 2013; 111: 1-10.
- Matsumoto S, Takahashi K, Iwakawa R, Matsuno Y, Nakanishi Y, Kohno T, et al. Frequent EGFR mutations in brain metastases of lung adenocarcinoma. Int J Cancer. 2006; 119: 1491-4.
- Sun M, Behrens C, Feng L, Ozburn N, Tang X, Yin G, et al. HER family receptor abnormalities in lung cancer brain metastases and corresponding primary tumors. Clin Cancer Res. 2009; 15: 4829-37.
- Han C, Zou H, Ma J, Zhou Y, Zhao J. Comparison of EGFR and KRAS status between primary non-small cell lung cancer and corresponding metastases: a systematic review and meta-analysis. Zhongguo Fei Ai Za Zhi. 2010; 13: 882-91.