Mini Review
J Stem Cell Res Transplant. 2014;1(1): 1003.
Protein Synthesis: More Than a House-Keeping Function in Hematopoietic Stem Cells
Rui Gao and Yan Liu*
Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN 46202, USA
*Corresponding author: Yan Liu, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN 46202, USA
Received: June 26, 2014; Accepted: July 30, 2014; Published: Aug 02, 2014
Hematopoietic stem cell (HSC) self-renewal is tightly regulated by both extrinsic signaling pathways and intrinsic regulators [1]. Adding to this list, in a recent issue of Nature, Signer et al. (2014) show that protein synthesis also plays a critical role in regulating hematopoietic stem cell self-renewal [2].
HSCs reside in a hypoxic niche in the bone marrow and majority of HSCs are in the quiescent state [3-4]. In addition, HSCs have low mitochondrial activity [5-6] and may have relatively lower rate of ribosome biogenesis. Thus, quiescent HSCs can be identified by using the Pyronin Y/Hoechst staining. Hoechst is an exclusive DNA dye while Pyronin Y reacts with both DNA and RNA. In the presence of Hoechst, Pyronin Y reaction with DNA is blocked, and Pyronin Y stains RNA only. When cells are stained first with Hoechst 33342 and then with Pyronin Y it is possible to distinguish DNA from RNA. Furthermore, quiescent cells, which are arrested in G0 phase, have lower level of RNA compared to active cells (G1 phase). The co-staining with the RNA dye Pyronin Y allows for the separation of G0 and G1 cell populations [7]. However, the protein synthesis rate in HSCs has not been determined.
It has been challengingto measure protein synthesis rate in vivo. Recently, Liu and colleagues developed a novelfluorogenic assay by using an alkyne analog of puromycin, O-propargyl-puromycin (OP-Puro), which can incorporate into newly synthesized peptides [8]. By injecting OP-Puro into mice, one can quantitate the protein synthesis rate in cells by using the flow cytometry analysis. Signer and colleagues utilized this novel method and found that HSCs and multi-potent progenitor cells (MPPs) synthesize less protein per hour compared to other hematopoietic cells, including common myeloid progenitors (CMPs), granulocyte and macrophage progenitors (GMPs) and terminally differentiated cells [2]. Interestingly, the reduced protein synthesis is not due to the differences in cell cycle status, cell size, ribosomal RNA content between HSCs and more mature cells.
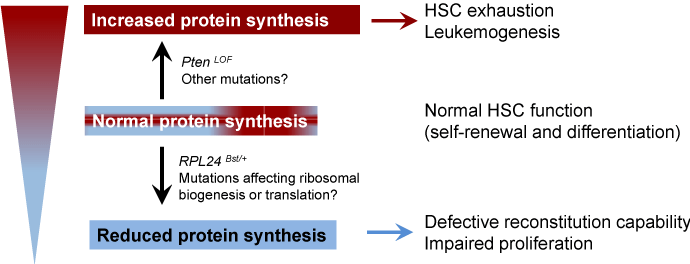
To examine whether the reducedprotein synthesis rate is important for HSC function, the authors utilized the RPL24Bst/+ mice, which harbors a loss-of-function mutation in the ribosomal protein gene RPL24. While these mice appear normal, there is a 30% decrease of protein synthesis rate [9]. RPL24 deficiency impairs both HSC self-renewal and proliferation potential in a cell-autonomous manner [2]. On the other hand, increasedprotein synthesis rate caused by Pten deletion depletes HSCs and promotes leukemogenesis [10]. Strikingly, crossing RPL24Bst/+ mice with Ptenfl/fl-Mx1-Cre mice restores HSC function and brings the proteinsynthesis rate to normal levels [2], indicating that a delicate balance in protein synthesis is critical for HSC function and tumor suppression (Figure 1). Mutations in ribosomal genes and other genes that are important for protein synthesis have been implicated inhuman diseases. While increased protein synthesis promotes development of certain cancers, defective ribosomal function causes ribosomopathies [11]. Ribosomopathies are human disorders of ribosome dysfunction, in which genetic abnormalities cause impaired ribosome biogenesis and function, resulting in specific clinical phenotypes [11].
Figure 1: A balanced protein synthesis rate is important for HSC function. Hematopoietic stem cells (HSCs) require appropriate level of protein synthesis to balance self-renewal and differentiation. Reduced protein synthesis impairs the ability for HSCs to self-renew, whereas increased protein synthesis depletes HSCs and promotes leukemogensis.
Impaired ribosomal function activates the p53 pathway, leading to cell cycle arrest and/or apoptosis [12]. However, the p53 pathway is not activated in RPL24Bst/+HSCs [2]. How protein synthesis is regulated in HSCs is largely unknown. It is possible that other mutations that affect ribosome biogenesis or translation will also reduce protein synthesis in HSCs. Indeed, reduced RPS19 or RPL11 expression in mouse erythroblast deregulates translation initiation of specific transcripts [13]. Investigation of whether other disease-associated ribosomal protein gene mutations affect protein synthesis will improve our understanding of how protein synthesis is regulated in HSCs.
It appears that different ribosomal proteins have distinct biological functions other than constituting ribosomes. In hematopoietic cells, RPS14haploinsufficiency and RPS19 mutations are associated with 5-q syndrome and Diamond-Blackfan Anemia (DBA), respectively [14-15]. The common mechanism of the two diseases is activation of the p53 pathway [11]. However, unlike the RPL24Bst/+ mutation, these mutations specifically affect erythroid cells while sparing other lineages [14-15]. There are many ribosomal protein gene mutations have been identified in DBA patients, including RPS24, RPS17, RPL35A, RPL5 and RPL11 [11]. Although these mutations share some common manifestations including anemia, macrocytosis, and absence of erythroid precursors, different ribosomal protein gene mutations display different phenotypes. For example, RPL5 gene mutations are associated with a higher frequency of physical abnormalities, including cleft lip and/or palate, whereas mutations in RPL11 had more isolated thumb abnormalities compared with patients with RPS19 mutations [16]. It will be interesting to investigate the distinct functions of different ribosomal proteins.
Overall, Signer and colleagues demonstrate that the protein synthesis in HSCs is tightly controlled. However, the exact mechanism underlying this process remains elusive. It is not clear why HSCs require lower rate of protein synthesis. Whether lower rate of protein synthesis is required for HSC self-renew awaits further investigation. Another interesting question is that whether there is an overall reduction of all protein species or only the synthesis of selective protein species is reduced in HSCs. Signer and colleagues report that HSCs and multi-potent progenitor cells (MPPs) have lower level of 4EBP1 phosphorylation compared to other hematopoietic cells, suggesting that the cap-dependent translation rate is low in HSCs and MPPs. How about cap-independent translation in HSCs and MPPs? Is cap-independent translation rate also low in HSCs? Reduced RPS19 and RPL11 expression in mouse erythroblasts impair the internal ribosomal entry site (IRES)–mediated translation of mRNAs [13]. It will be interesting to investigate whether HSCs also shut down IRES-dependent translation of mRNAs.
While it is clear that protein synthesis is important for HSC function, many questions remain unanswered. What are the links between hypoxic environment and mitochondrial activity? What is the relationship between hypoxic environments and stem cells with respect to energy production and therefore protein production? Why would decreased protein production be important for a stem cell (or just HSCs)? What is the link between potency and protein production? Further research should be directed to address these important questions and uncover the full regulatory network involved in HSC regulation. Hematopoietic stem cell is the best characterized somatic stem cell so far and has become a model for stem cell research [1]. It is highly possible that a balanced protein synthesis rate is essential for the maintenance of other stem cells as well.
References
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012; 481: 457-462.
- Signer RAJ, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014; 509: 49-54.
- Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol. 2011; 12: 643-655.
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008; 135: 1118-1129.
- Simsek T, Kocabas F, Zheng J, DeBerardinis RJ, Mahmoud AI. The Distinct Metabolic Profile of Hematopoietic Stem Cells Reflects Their Location in a Hypoxic Niche. Cell Stem Cell. 2010; 7: 380-390.
- Suda, T, Takubo K, Semenza, Gregg L. Metabolic Regulation of Hematopoietic Stem Cells in the Hypoxic Niche. Cell Stem Cell. 2011; 9: 298-310.
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009; 4: 37-48.
- Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012; 109: 413-418.
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008; 456: 971-975.
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006; 441: 475-482.
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood 2010; 115: 3196-3205.
- Zhang Y, Lu H. Signaling to p53: Ribosomal Proteins Find Their Way. Cancer Cell. 2009; 16: 369-377.
- Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood 2012; 119: 262-272.
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, et al. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007; 109: 980-986.
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008; 451: 335-339.
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, et al. Ribosomal Protein L5 and L11 Mutations Are Associated with Cleft Palate and Abnormal Thumbs in Diamond-Blackfan Anemia Patients. Am J Hum Genet. 2008; 83: 769-780.