Special Article – Surgery Case Reports
Austin J Surg. 2019; 6(4): 1167.
EGFR Blockade as Effective Therapy in BRAF and EGFR Mutated Metastatic Colorectal Cancer: Learning from a Clinical Case
Ghiglione L¹, Esposito F¹, Pagés M², Jares P³ and Maurel J¹*
¹Department of Medical Oncology, IDIBAPS, Universitat de Barcelona c/ Villarroel, Spain
²Department of Radiology, Centre de Diagnòstic per la Imatge Clínic (CDIC), Spain
³Department of Anatomic Pathology, Hospital Clinic of Barcelona, University of Barcelona, Barcelona, Spain
*Corresponding author: Joan Maurel, Medical Oncology Department, Hospital Clínic, Translational Genomics and Targeted Therapeutics in Solid Tumors Group, IDIBAPS, Universitat de Barcelona, c/ Villarroel, Spain
Received: December 31, 2018; Accepted: February 08, 2019; Published: February 15, 2019
Abstract
Metastatic colorectal cancer patients affected of BRAF mutations are associated with poor prognosis. The co-association with EGFR mutation in the tyrosine kinase domain is quite low. We report a case of an EGFR and BRAF mutated patient and its unexpected outcome. A search through the literature for EGFR mutated and information about its response to EGFR blockade was perform. Likewise, we reviewed our own cohort of patients. The case presented and our analysis reinforces the importance of using next generation sequencing to discover unusual mutations tributary of anti-EGFR therapy.
Keywords: EGFR mutated; BRAF mutated; Metastatic colorectal cancer; NGS; Cetuximab
Introduction
Colorectal Cancer (CRC) is the second most commonly diagnosed cancer and one of the leading causes of cancer deaths worldwide. Approximately 50% of metastatic CRC harbour RAS mutations and predicts anti-EGFR compounds (cetuximab and panitumumab) resistance [1]. Roughly, 5% of mCRC have microsatellite instability (MSI) and these patients are sensitive to immune checkpoint blockade [2]. BRAF mutation is associated with poor prognosis [3] but the role is still controversial as a predictive marker of anti-EGFR therapy [4- 6]. Next generation sequencing (NGS) allows to evaluate targeteddriven genes that are rarely mutated, fussed or amplified in mCRC (HER2, EGFR, MET and FGFR) and there use in clinical setting is currently increasing.
We present a case of a BRAF mutant patient mCRC, who experienced a long-term complete response with FOLFOX plus cetuximab in the POSIBA trial (NCT01276379).
Case Presentation
A 44-year-old Caucasian woman was referred due to abdominal pain and anaemia on April 2012. Blood test revealed Hb 11 gr/dL and CEA 9.5 ng/mL (reference range <5 ng/mL). An abdominal Computed Tomography (CT) scan described a mass on the left side colon and pathologic retroperitoneal lymph nodes. Colonoscopy reports a left colonic lesion. While preparation for surgical intervention, she presented symptoms of intestinal obstruction and a subtotal colectomy was done on April 27, 2012. Pathologic specimen shows a high-grade mucinous adenocarcinoma, pT3N2 (33/66), KRAS (exon 2) Wild Type (WT). Postoperative thoracoabdominal CT described pathologic mesenteric, retroperitoneal and mediastinal lymph nodes. PET-CT showed lymph nodes with increased metabolic activity in the same territory depicted on the CT. Because of the unusual presentation with thoracic lymph nodes, a bronchoscopy with biopsy was performed reporting adenocarcinoma (CAM5.2+). Patient was included in the clinical trial POSIBA on July 2012 and received FOLFOX6m plus cetuximab 500 mg/m2 every 2 weeks for 12 cycles followed by cetuximab monotherapy discontinued because of grade 3 cutaneous toxicity in 17th cycle on March 2013. A 3-month reevaluation CT showed a complete response which is maintained until today on June 2018 (Figure 1). Extended RAS and BRAF mutations were done per protocol on 10/2015 and a BRAF (V600E) mutation was detected.
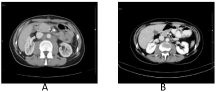
Figure 1: A. Scanner, May 2012, before treatment B. Scanner, June 2013,
after.
In April 2016 we further analysed the sample obtained on surgery due to its unusual evolution and the availability of NGS in our Center. Tumour Mismatch-Repair Deficiency (MMR) status determined by Immunohistochemical (IHC) was informed as Microsatellite Stable (MSS). NGS showed mutations in SMAD4 (R361C), EGFR (E749K) and BRAF (V600E) with an allele frequency of 65%, 18% and 18%, respectively.
Discussion
There are two major domains with EGFR mutations in CRC. First domain is located in the ligand-binding-domain at exon 12 and appeared related to cetuximab and panitumumab acquired resistance [7]. Second domain is located in the Tyrosine Kinase Domain (TKD) (exons 18-21) and potentially confers cetuximab sensitivity [8]. The EGFR mutation (E749K) in the TKD has been previously described in CRC [9]. Silico computer-based modelling suggests a potential pathogenic role. The incidence of EGFR mutations in the TKD in CRC is quite low (25/2658; 0.9%) in 9 Asiatic and non-Asiatic cohorts; range (0.03-0.3%) [9-18] (Figure 2). We have reviewed our prospective consecutive cohort of 167 mCRC patients (Table 1) evaluated with NGS from February 2016 to June 2018 and only one patient (0.6% frequency) have an EGFR mutation. This patient has an EGFR mutation located on exon 21 L858R (9% allele frequency) and concomitant mutations in PIK3CA (G542L), BRAF (V600E) and p53 (R249M and G245S). This mutation is observed in 40% of EGFR mutations in non-small cell lung cancer and was previously described in CRC [10]. Complete responses with anti-EGFR compounds have been described more frequently in RAS and BRAF WT (double WT) patients with EGFR mutations (3/4; 75%), than in double WT without EGFR mutations (4/61; 6%) [19]. To our knowledge, long-term complete responses with chemotherapy associated with cetuximab in mCRC patients with concomitant presence of BRAF and EGFR mutations has not been previously reported.
MSS
MSI-H
TOTAL N=167
N=158
94,61%
N=9
5,39%
RAS
86
54,43%
2
22,22%
p53
107
67,72%
3
33,33%
BRAF
19
12,03%
4
44,44%
PI3K
21
13,29%
2
22,22%
PTEN
3
1,90%
1
11,11%
FBXW7
14
8,86%
1
11,11%
SMAD4
20
12,66%
2
22,22%
AKT1
2
1,27%
0
0,00%
MET
1
0,63%
1
11,11%
ERB4
1
0,63%
0
0,00%
MAP2K1
2
1,27%
0
0,00%
DDR2
0
0,00%
1
11,11%
STK11
1
0,63%
0
0,00%
MAPK2
1
0,63%
0
0,00%
ERB2
1
0,63%
1
11,11%
CTNNB1
2
1,27%
0
0,00%
EGFR
0
0,00%
1
11,11%
TOTAL MUTATIONS
281
19
Abbreviations: CAM5.2: Cytokeratin CAM 5.2; CEA: Carcinoembryonic Antigen; CRC: Colorectal Cancer; CT: Computed Tomography; EGFR: Epidermal Growth Factor Receptor; FGFR: Fibroblast Growth Factor Receptor; FOLFOX: 5-Fluorouracil and Oxaliplatin Chemotherapy; Hb: Haemoglobin; HER2: Human Epidermal Growth Factor Receptor 2; IHC: Immunohistochemical; mCRC: Metastatic Colorectal Cancer; MET: Tyrosine-Protein Kinase Met; MMR: Mismatch-Repair Deficiency; MSI-H: Microsatellite Instability-High; MSS; Microsatellite Stability; NGS: Next Generation Sequencing; p53: Tumour Protein p53; PET-CT: Positron Emission Tomography-Computed Tomography; PIK3CA: Catalytic Subunit of Phosphatidylinositol 3-Kinase; RAF: Rapidly Accelerated Fibrosarcoma; Serine/threonine-specific protein kinase; RAS: Oncogene of Rat Sarcoma Protein; SMAD4: Similarity Mothers Against Decapentaplegic to the Drosophila Gene; TKD: Tyrosine Kinase Domain; WT: Wild Type
Table 1: NGS mCRC analysis of patients from February 2016 to June 2018.
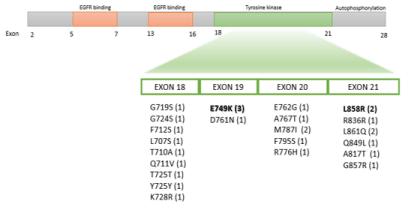
Figure 2: EGFR mutations in kinase domain described in mCCR.
The mutations described in the literature and their frequency in breakets. In bolde our patients findings.
Conclusion
We presented the case of a 44-year-old woman affected by mutations on EGFR, BRAF and SMAD genes, who experienced longterm complete response with FOLFOX plus cetuximab. The result was impressive and reinforce the importance of NGS, to discover unusual mutations tributary of anti-EGFR therapy.
References
- Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011; 29: 2011-2019.
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015; 372: 2509-2520.
- Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014; 20: 5322-5330.
- Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015; 112: 1888-1894.
- Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015; 51: 587-594.
- van Brummelen EMJ, de Boer A, Beijnen JH, Schellens JHM. BRAF Mutations as Predictive Biomarker for Response to Anti-EGFR Monoclonal Antibodies. Oncologist. 2017; 22: 864-872.
- Arena S, Bellosillo B, Siravegna G, Martínez A, Cañadas I, Lazzari L, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin Cancer Res. 2015; 21: 2157-2166.
- Cho J, Bass AJ, Lawrence MS, Cibulskis K, Cho A, Lee S-N, et al. Colon cancer-derived oncogenic EGFR G724S mutant identified by whole genome sequence analysis is dependent on asymmetric dimerization and sensitive to cetuximab. Mol Cancer. 2014; 13: 141.
- Nagahara H, Mimori K, Ohta M, Utsunomiya T, Inoue H, Barnard GF, et al. Somatic mutations of epidermal growth factor receptor in colorectal carcinoma. Clin Cancer Res. 2005; 11: 1368-1371.
- Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018; 33: 125-136e3.
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23: 703-713.
- D’Haene N, Fontanges Q, De Nève N, Blanchard O, Melendez B, Delos M, et al. Clinical application of targeted next-generation sequencing for colorectal cancer patients: a multicentric Belgian experience. Oncotarget. 2018; 9: 20761-20768.
- Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004; 351: 2883.
- Oh BY, Lee RA, Chung SS, Kim KH. Epidermal growth factor receptor mutations in colorectal cancer patients. J Korean Soc Coloproctol. 2011; 27: 127-132.
- Ogino S, Meyerhardt JA, Cantor M, Brahmandam M, Clark JW, Namgyal C, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005; 11: 6650-6656.
- Metzger B, Chambeau L, Begon DY, Faber C, Kayser J, Berchem G, et al. The human epidermal growth factor receptor (EGFR) gene in European patients with advanced colorectal cancer harbors infrequent mutations in its tyrosine kinase domain. BMC Med Genet. 2011; 12: 144.
- Yunxia Z, Jun C, Guanshan Z, Yachao L, Xueke Z, Jin L. Mutations in epidermal growth factor receptor and K-ras in Chinese patients with colorectal cancer. BMC Med Genet. 2010; 11: 34.
- Moroni M, Veronese S, Benvenuti S, Marrapese G, Sartore-Bianchi A, Di Nicolantonio F, et al. Gene copy number for Epidermal Growth Factor Receptor (EGFR) and clinical response to anti EGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005; 6: 279-286.
- Lupini L, Bassi C, Mlcochova J, Musa G, Russo M, Vychytilova-Faltejskova P, et al. Prediction of response to anti-EGFR antibody-based therapies by multigene sequencing in colorectal cancer patients. BMC Cancer. 2015; 15: 808.