Research Article
Austin J Surg. 2019; 6(8): 1179.
DDX49 May be a Novel Biomarker and Therapeutic Target of Lymph Node Metastases in Lung Cancer
Lian X¹*, Xing Z²*, Sun G¹, Bing X¹, Peng C¹, Liao M² and Zhang Z²
¹Department of Tumor Blood, Jiangjin Central Hospital, China
²Department of Surgery, Cancer Center and Daping Hospital, Amry Medical University, China
*Corresponding author: Zhimin Zhang, Cancer Center, Daping Hospital and Research Institute of Surgery, Army Medical University, China
Received: February 25, 2019; Accepted: April 16, 2019; Published: April 23, 2019
Abstract
The identification of lymph node metastases is important for the diagnosis, treatment and prognosis of patients with lung cancer. We found DDX49 was associated with the lymph node metastases in lung cancer by the Akt/β-catenin pathway. Transcriptome sequencing, bioinformatics analysis, quantitative RTPCR, Cell Transfection and The Cancer Genome Atlas (TCGA) dataset were used to identify DDX49 responsible for lymph node metastases. A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were used to explore the possible molecular mechanism in cell experimental. The DDX49 gene was correlated significantly with lymph node metastases of lung cancer. The know down of DDX49 inhibited the cell proliferation and migration in PC-9 and H460 cells. The mechanism research found down expression of DDX49 decreased the Akt/β-catenin pathway in lung cancer cell. In vivo experiments showed that DDX49 promoted the proliferation and metastases of lung cancer cells by increasing the Akt/β-catenin pathway. These findings suggested that DDX49 may be useful as a novel biomarker of lymph node metastases and a therapeutic target in lung cancer therapy.
Keywords: Lymph node metastases; Lung cancer; DDX49; Predictor; Therapeutic target
Introduction
Now, the highest occurrence and fatality rate was lung cancer in the world. The accurate diagnosis, especially the diagnosis of distant metastases is one of the important factors to improve the prognosis of lung cancer therapy. The first diagnosis of lung cancer is often with distant metastases, especially lymph node metastases, so patients have poor 5-year survival rates. Lymph node metastases has a higher incidence rate and is not only one of the most important factors determining the cancer stage but also an important indicator of treatment options in lung cancer [1]. The current diagnosis for lymph node metastases of lung cancer has not a precise maker. Only less than 80% accuracy in detecting lymph node metastases is achieved by enhanced CT, which is the first choice of diagnostic methods [2]. The approximately 20% of lymph node metastases remain undetected by the routinely methods. Patients with undetected T3 metastases will not benefit from radical surgical resection and may experience postoperative recurrence [3]. In addition, this is not a precise target of lymph node metastases. Therefore, it is essential to look for a new marker for individualized treatment options to accurately consider the stage of the cancer to decrease adverse events in patients without necessary treatment.
It was an important step towards being able to implement individualized therapy that the identification of molecular characteristics that could predict the existence of undetected lymph node metastases in lung cancer. In our research, we aimed to identify DDX49 that accurately predict lymph node metastases of lung cancer using transcriptome sequencing, bioinformatics analysis, quantitative RT-PCR, cell transfection mothed, and 188 samples from The Cancer Genome Atlas (TCGA) dataset. We obtained a new predictor DDX49. In addition, we found that DDX49 was associated with cell proliferation and migration in PC-9 and H460 cells. The mechanism research found, DDX49 regulated cell proliferation and migration by the Akt/β-catenin pathway using the Lentivirus-knockdown assay, biological indicators and in vivo experiments. The identification of DDX49 could help diagnose a subgroup of lung cancer patients with lymph node metastases, who are likely candidates for individualized therapy.
Methods and Materials
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM) and Fetal Bovine Serum (FBS) were obtained from Invitrogen (Grand Island, NY, USA). Penicillin, Streptomycin and Dimethyl Sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Antibodies against DDX49, P-Akt and Akt were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against DDX49, β-tubulin, and Horseradish Peroxidase (HRP)-conjugated antimouse and anti-rabbit IgG antibodies were from Abcam (Cambridge, MA, USA). Transwell® chambers (Corning, Tewksbury, MA, USA), crystal violet, loading buffer and RIPA assay were purchased from Beyotime Corporation.
TCGA dataset
A preprocessed expression matrix of gene-level RSEM values from 188 cases of lung cancer originally documented in TCGA datasets (Supplemental Table 1) Clinical characteristics of lung cancer patients (https://tcga.xenahubs.net/download/TCGA.LUNG. sampleMap/AgilentG4502A_07_3.gz). Clinical information of this TCGA cohort such as age, gender, histology, TNM stage and clinical stage were also obtained from UCSC (https://tcga.xenahubs.net/ download/TCGA.LUNG.sampleMap/LUNG_clinicalMatrix.g).
RNA preparation and qualification and transcriptome sequencing
Total RNA was isolated with RNeasy plus Mini Kit (Catalog no. 74134, Qiagen, Duesseldorf, German). RNA concentration and integrity were detected by the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®Ultra™ RNA Library Prep Kit for Illumina®(NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on ac Bot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and paired-end reads were generated. Index of the reference genome was built using Bowtie v2.2.3 and. Reads were blasted to the reference genome using TopHat v2.0.12. The expression level of each gene was conducted based on RNA-seq was measured as numbers of Reads Per Kilo base of Exon Region in a Gene Per Million Mapped Reads (RPKM). Differential expression analysis of two samples was performed using the DEGseq (2010) R package. P value was adjusted using q value (19). FDR <0.05 and |log2Ratio|=1 found by DESeq was set as the threshold for significantly differential expression.
Data analysis
Clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Then, Q20, Q30 and GC content the clean data were assessed. The clean data with high quality was the basis for all the following analyses.
Pathway enrichment analysis
A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted using the Functional Annotation tool in DAVID Bioinformatics Resources. Gene sets linked to positive lymph node metastasis of lung cancer or negative were used as input in DAVID for the mining of the functional relevance of the lymph node metastasis.
Real-time PCR
The expression levels of the 58 genes were validated by real time RT-PCR. The cDNA was synthesized with RevertAid Fist Strand cDNA Synthesis Kit (ThermoFisher, United States, Category Number: #K1622). Primers purchased from Bioligo Inc. (Shanghai China). Real time quantitative RT-PCR was performed with the SYBR Green PCR method using Fast SYBRTM Green Master Mix (ThermoFisher, United States, Category Number: 4385610) with CFX96TM Real-Time System (Bio-Rad, United States). GAPDH was used as endogenous normalization controls. 2-==Ct method was used to calculate the expression levels. Real-time PCR was performed in duplicate in three dependent experiments.
Cell culture
Human lung cancer cells lines, PC-9 and H460, were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cells were cultured in MEM medium containing 10% FBS and 50 mg/mL penicillin/streptomycin, in a 5% CO2, 37°C humidified incubator.
Transfection
shRNA-DDX49 (shDDX49) and shRNA-con (shCON) plasmid were purchased from Genechem Corporation and used according to manufacturer’s instructions with a Silencer shRNA construction kit (Qiagen, Valencia, CA). Cells were incubated with shRNA transfection complexes at 40% confluence. shRNA incubations were performed with RNAi Fect reagents (Qiagen). Forty-eight hours after transfection, cells were treated.
Western blot analysis
Equal amounts of proteins were separated on SDS-polyacrylamide gels, transferred to PVDF membranes, and then blocked with 5% nonfat dry milk in TBST for 1 h at room temperature. Next, membranes were incubated with primary antibodies overnight at 4°C, followed by 1 h incubation at room temperature with horseradish peroxidaseconjugated secondary antibodies. Finally, blots were imaged using chemiluminescent staining reagents. The stained protein bands were visualized with BioMax-Light film (Eastman Kodak Co., Rochester, NY, USA), and the staining intensities of the various protein bands were obtained using the Gel Doc 2000 apparatus and software (Quantity One, Bio-Rad; Hercules, CA, USA).
Scratch assay
Cells were plated in 6-well culture plates and incubated overnight to reach a density of 60%–70%. Cell monolayers were then scratched with a 100-μL yellow pipette tip and washed with PBS three times to remove detached cells. The wounded areas were imaged using an Olympus microscope and marked. The indicated doses of palbociclib, celecoxib or LPS were applied to culture cells for 48 hours at 37°C in 5% CO2. The culture medium was removed, and the same areas were imaged again to observe the wound gap.
Transwell assay
The Matrigel invasion assay was carried out using 8-μm pore size transwell filters (Corning, 6.5 mm diameter). The cell-culture inserts were coated with 5 μl pure Matrigel (Sigma, St. Louis, MO) and placed in a 24-well plate. Cells were detached and single-cell suspensions were placed at 1×105 cells per well into the upper chamber in 0.1 ml of serum-free medium. In the lower chamber, DMEM or Eagle’s minimal essential medium supplemented with 10% FBS was placed as a chemo-attractant. Celecoxib was added at constant concentration to both the upper and the lower chambers. After 18 h of incubation in 5%CO2 at 37°C, the filters were fixed with 90% ethanol for 30 min and stained with crystal violet. Cells from the upper surface of the chamber were removed with gentle swabbing and the invasive cells on the lower surface of the filters were examined by bright field microscopy.
In vivo experiment
Sixteen BALB/C nude mice (female, aged 6–8 weeks) were obtained from the Daping Hospital of the Third Military Medical University Laboratory Animal Co Ltd. (Chongqing, People’s Republic of China), divided into 4 groups (each group contained 4 mice) and housed in a pathogen-free environment under controlled conditions (temperature 20°C–26°C, humidity 40%–70%, light–dark cycle 12–12 hours). The mice were injected subcutaneously with 3×106 H460 cells that had been stably transfected with a plasmid containing DDX49 mutation (shDDX49) or cells that had received a vehicle (shCON) suspended in Phosphate-Buffered Saline (PBS). Tumor diameter was measured twice a week using calipers, and tumor volume was calculated as follows: ab2/2 mm3, where is the length and is the width of the tumor. The protocol was reviewed by the Ethics Committee of the Daping Hospital and Research Institute of the Third Military Medical University. The animal experiments were performed in accordance with the Guidelines for the Accommodation and Care of Laboratory Animals at the Daping Hospital and Research Institute of the Third Military Medical University. Mice that developed tumors reaching approximately 100 mm3 in size were randomized into four groups with ten mice in each group: shCON and shDDX49.
Patients
10 specimens used to transcriptome sequencing and bioinformatics analysis were obtained from surgically resected primary tissue in patients with lung cancer treated at Daping Hospital of the Amry Medical University (China) and were frozen and kept in liquid nitrogen (Supplemental Table 2). Clinical characteristics of lung cancer patients. The study protocol was approved by the Ethics Committee of the Daping Hospital and Research Institute, Third Military Medical University.
Statistical analysis
58 significantly differentially expressed genes were analyzed to identify candidates potentially predictive for lymph node metastasis using univariate logistic regression. 1 gene was found associated with lymph node metastasis with crude P value less than 0.05. All other statistical analyses were performed using SPSS 17.0 (IBM SPSS, Chicago, IL, USA). All tests were bilateral, and P < 0.05 was considered statistically significant.
Results
DDX49 gene were associated with lymph node metastases of lung cancer
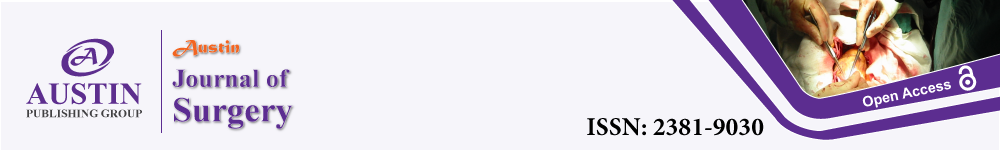
To research the molecular nature of lymph node metastases in lung cancer, a sequential approach was used to select genes associated with lymph node metastases (Figure 1A). by selecting [1] genes differentially expressed in positive versus negative lymph node metastases; [2] genes associated with lung cancer, [3] genes was associated with cell proliferation in lung cancer cell, [4] genes were further identified in TCGA data of 188 sample Transcriptome Sequencing results (Figure 1A), with a crude P value of less than 0.05.
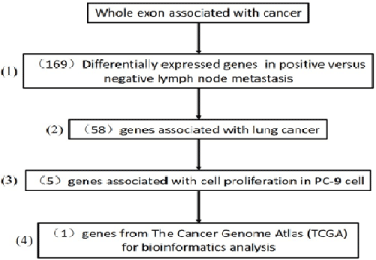
Figure 1: TCGA data of 188 sample Transcriptome Sequencing results.
Effects of DDX49 on lung cancer cell growth and migration
Then, we investigated the role of DDX49 in cell proliferation and migration in lung cancer cells by a lentivirus-knockdown assay. Downregulation of DDX49 in PC-6 and H460 cells, as shown in (Figure 2A), substantially decreased cell growth and cell migration, all of which were increased following cisplatin treatment (Figure 2B-D).
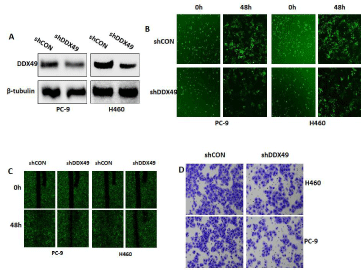
Figure 2: Downregulation of DDX49 in PC-6 and H460 cells.
Figure 3: The role of DDX49 in cell proliferation and migration in lung cancer
cells.
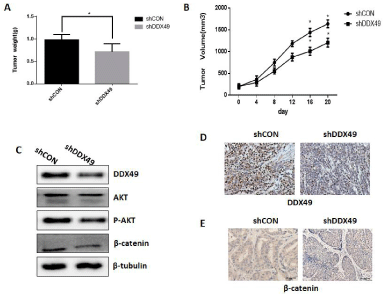
Figure 4: The downregulation of DDX49 significantly decreased tumor
growth.
Moreover, we applied immunoblotting to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with lymph node metastases of lung cancer, with a corrected P< 0.0001 and expression in more than half of the samples (Supplemental Table 3). KEGG pathway enrichment analysis of all genes selected from all differentially expressed genes showed that the PI3K/Akt and Wnt/β- catenin pathways may play key roles in lymph node metastases (Figure 3A). We demonstrated that DDX49 downregulation decreased Akt and β-catenin activation (Figure 3B). These findings indicated that decreases in the activation of Akt/β-catenin signaling by DDX49 knockdown may play a key role in inhibiting cell proliferation and migration in vitro.
Downregulation of DDX49 inhibited tumor growth in vivo
We studied the impact of DDX49 on cell growth in lung cancer cells by injecting H460 cells containing either a control (shCON) or DDX49-downregulation vector (shDDX49) into nude mice in vivo. We observed that downregulation of DDX49 significantly decreased tumor growth in vivo (Figure 4A-B). Consistent with observations in vitro, shDDX49 decreased DDX49 expression in vivo (Figures 4C and D), and DDX49 downregulation decreased Akt and β-catenin activation (Figures 4C and E). These findings indicated that DDX49 downregulation decreases the activation of Akt/β-catenin signaling and may play a key role in inhibiting cell proliferation and migration in vivo.
Discussion
Accurate diagnosis of lymph node metastases is a key factor in individualized lung cancer therapy and 5-year survival. Current examination methods for accurately assessing potential lymph node metastases require further exploration. In the present study, we integrated transcriptome sequencing data of lymph node metastases in lung cancer and an independent sample from TCGA data to identify DDX49 gene and validated the biological function in vitro and in vivo. Furthermore, we demonstrated, for the first time, that DDX49 promoted lung cancer cell growth and migration by increasing the Akt/β-catenin pathway. Taken together, these findings suggested that DDX49 may be useful as a novel biomarker of lymph node metastases and a therapeutic target in lung cancer therapy.
Our selection of DDX49 gene in the microarray detection set, and the patterns of gene expression found on microarray analysis were also validated by RT-PCR. Our results were validated in one independent cohorts of 188 patients from TCGA dataset. Thus, we believe that the DDX49 we obtained using the three factors is reliable.
The identification of DDX49 genes that can predict lymph node metastases in patients with lung cancer may reveal a new target of therapy for lung cancer with lymph node or distant metastases. DDX49 is one such uncharacterized RNA helicase [4], which has been implicated in viral infections and breast cancers in high throughput screens, suggesting an important physiological role [5]. We found that downregulation of DDX49 in lung cancer cells significantly suppressed NSCLC (non-small cell lung cancer) cell growth and migration. Sharad Awasthi also proposed a similar conclusion: DDX49 displays a robust ATPase and RNA helicase activity and it is involved in the export of poly (A)+ mRNAs to cytoplasm, DDX49 affects cell proliferation and its aberrant expression might have oncogenic potential [6]. Identifying the associated KEGG pathways, we found that these genes were more likely involved in the PI3K/ Akt pathway, cell and metabolic pathways, and focal adhesion. Furthermore, we found that DDX49 mediated by the Akt/β-catenin pathway promoted NSCLC cell growth and metastases. Growing evidence has shown that the Akt/β-catenin pathway plays a key role in lung cancer cells [7,8]. Together, DDX49 plays an important role through the Akt/β-catenin pathway in increasing cell growth and metastases. DDX49 may be a new target for therapy of lung cancer with lymph node or distant metastases.
DDX49 was a novel predictor for diagnosing lymph node metastases of lung cancer, thus, the marker can be used to improve or even replace enhanced CT for the diagnosis of lymph node metastases. Moreover, inhibition of DDX49 can significantly inhibit cell proliferation and metastases by the Akt/β-catenin pathway in vitro and in vivo. DDX49 may be useful as a novel biomarker of lymph node metastases and as a therapeutic target in lung cancer.
Acknowledgement
This work was supported by grants from the National Natural Science Foundation of China (NSFC) No. 81802292 to Zhimin Zhang.
Disclosure statement: This study was carried out after approval by the Ethics Committee of the Daping Hospital and Research Institute of Surgery, Army Medical University and obtaining informed consent from all subjects.
Competing Interests: No potential conflicts of interest were disclosed.
References
- Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, et al. Non-Small Cell Lung Cancer, Version 6.2015. Journal of the National Comprehensive Cancer Network : JNCCN. 2015; 13: 515-524.
- Zhou H, Liu JK, Chen SX, Xiong Z, Lin GQ, Zhou ML, et al. Lymphatic microvessel density combined with CT used in the diagnosis of mediastinal and hilar lymph node metastasis of non-small cell lung cancer. Archives of medical research. 2012; 43: 132-138.
- Naruke T, Goya T, Tsuchiya R, Suemasu K. The importance of surgery to non-small cell carcinoma of lung with mediastinal lymph node metastasis. The Annals of thoracic surgery. 1988; 46: 603-610.
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006; 367: 17-37.
- Apostolou P, Toloudi M, Papasotiriou I. Identification of genes involved in breast cancer and breast cancer stem cells. Breast Cancer (Dove Med Press). 2015; 7: 183-191.
- Awasthi S, Verma M, Mahesh A, MI KK, Govindaraju G, Rajavelu A, et al. DDX49 is an RNA helicase that affects translation by regulating mRNA export and the levels of pre-ribosomal RNA. Nucleic acids research. 2018; 46: 6304- 6317.
- Liang CH, Chiu SY, Hsu IL, Wu YY, Tsai YT, Ke JY, et al. alpha-Catulin drives metastasis by activating ILK and driving an alphavbeta3 integrin signaling axis. Cancer research. 2013; 73: 428-438.
- Zhao M, Xu P, Liu Z, Zhen Y, Chen Y, Liu Y, et al. Dual roles of miR-374a by modulated c-Jun respectively targets CCND1-inducing PI3K/AKT signal and PTEN-suppressing Wnt/beta-catenin signaling in non-small-cell lung cancer. Cell death & disease. 2018; 9: 78.