
Research Article
Thromb Haemost Res. 2017; 1(1): 1005.
Lipoprotein (a) and Interleukin-6 Genetic Polymorphisms Interactions Associated with Cardiovascular Events in the Mexican Population
Lopez-Salazar LI¹, Hernández-Tobias EA², Noris G³, Santana C³, Meraz-Ríos MA4, Quiroz-Jimenez N¹, Brooks D5, Calderón-Aranda ES1, Majluf A6 and Gomez R1*
¹Departamento de Toxicología, Cinvestav-IPN, Mexico
²Universidad Autónoma de Nuevo León, Facultad de Salud Pública y Nutrición, Mexico
³Laboratorio Biología Molecular Diagnóstica, Mexico
4Departamento de Biomedicina Molecular, Cinvestav- IPN, Mexico
5Anthropology Department, University of Pensylvania, USA
6Unidad de Investigación Médica en Trombosis, Hemostasia y Aterogénesis, Mexico
*Corresponding author: Gomez R, Departamento de Toxicología, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional. Av. Instituto Politécnico Nacional 2508, Col. San Pedro Zacatenco, 07360, Mexico City, México
Received: June 16, 2017; Accepted: August 22, 2017; Published: September 04, 2017
Abstract
Cardiovascular diseases are complex conditions whose more prominent clinical manifestations more prominent are coronary arterial disease, myocardial infarction, and stroke, the principal causes of death worldwide. Although different risk factors have been related to their development, heritability is one of the most relevant predictors. Thus, the exploration of different loci using genetic polymorphisms is the most used strategy to find risk biomarkers. Genes related to lipid metabolism, pro-thrombotic and inflammation processes have been the most studied. Overall, of genetic association studies research genes of specific biological processes. Nevertheless, the interaction among genes involved in different process is the best approaches to find genotypephenotype associations, as shown in the genome-wide association studies. Likewise, to avoid bias related to the ethnic background genomic and ancestral controls should be included. Herein, lipoprotein (a) and interleukin 6 genetic polymorphisms were analysed. Our findings suggest an interaction between these genetic polymorphisms that could contribute to the development of cardiovascular diseases. Additionally, the inclusion of the genomic and ancestral controls evidenced an interethnic variation in the studied populations, reflecting the influence of demographic processes in the search for risk biomarkers. Finally, the present study reinforces the contribution of the polymorphisms on lipoprotein (a) gene in the incidence of cardiovascular events.
Keywords: Lipoprotein (a); Interleukin-6; Genetic polymorphisms; Atherothrombosis; Mexico
Abbreviations
AGC: Ancestral Genome Control; BMD: Bone Mineral Density; BMI: Body Mass Index; CEPH: Centre d’Etude du Polymorphisme Humain; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; CI: Confidence Interval; CINVESTAV: Center for Research and Advanced Studies; CNV: Copy Number Variable; CVDs: Cardiovascular Diseases; DNA: Deoxyribonucleic Acid; GAS: Genetic Association Studies; GC: Genome Control; GWAS: Genome-Wide Association Studies; HbA1cglycated: Haemoglobin A1c; HDL: High Density Lipoprotein; HWE: Hardy-Weinberg Equilibrium; IL-6: Interleukin-6; IL6: Interleukin-6 Gene; IMSS: Instituto Mexicano del Seguro Social; IC: Interquartile Range; k: Number of Subpopulations; kcal/ day: Kilocalories Per Day; KIV-2: Kringle-type 2; LD: Linkage Disequilibrium; LDL: Low Density Lipoprotein; Lp(a): Lipoprotein (a); LPA: Lipoprotein(a)gene; MACs: Multi-Allelic Combinations; MAF: Minor Allele Frequency; MDS: Multidimensional Scaling; METs: Metabolic Equivalents; mg/dL: Milligrams Per Decilitre; MI: Myocardial Infarction; min/week: Minutes Per Week; mmHg: Millimetres of Mercury; MXL: Mexican ancestry in Los Angeles, California; OR: Odds Ratio; PCR: Polymerase Chain Reaction; PEL: Peruvian in Lima, Peru; SEM: Standard Error of the Mean; SNP: Single Nucleotide Polymorphisms; STREGA: Strengthening the Reporting of Genetic Association Studies; TSI: Toscani in Italy; WES: Whole Exome Studies.
Introduction
Cardiovascular Diseases (CVDs) occupy the higher mortality rates worldwide [1]. These complex conditions are related to the lifestyle, behavioural risk factors (i.e., tobacco use, alcohol consumption, among others) and to genetic predisposition [2]. Heritability is a strong predictor of susceptibility and a fruitful tool to identify risk biomarkers. Genome-Wide Association Studies (GWAS) and more recently Whole Exome Studies (WES) support that hypercoagulability, hypofibrinolysis, oxidative and inflammatory processes and lipid metabolism are keystones in CVDs development [3,4]. Lipoprotein(a) -Lp(a)-, a homologous protein to plasminogen, is involved in the vascular remodelling and tissue repairing [5]. Further, Lp(a) has a critical role in pro-inflammatory and pro-atherogenic oxidised phospholipids formation, being related to atherosclerosis and thrombosis risk, and its subsequent clinical manifestations [6,7]. Of note the high heritability of Lp(a) plasma concentrations associated with genetic variants in LPA gene, which has been broadly studied albeit in Mexico this kind of studies are scanty [5].
Pro-inflammatory cytokines have been related to the clinical prognosis of CVDs [8]. Particularly, interleukin-6 (IL-6) increases platelet count and aggregation, participates in endothelial cells activation and consequently induces a prothrombotic state [9]. Although different Genetic Association Studies (GAS) have explored the participation of IL-6 as a risk marker, the results have been inconclusive [10,11]. The discrepant results among different populations could be related to the complex genetic architecture of multi-ethnic populations where some biochemical determinations could be variables depending on the ethnicity [12]. Moreover, the best approach to identify promising biomarkers involves the study of multiple genes in different biological processes as well as the interactions among them [13].
The present study evaluates the allelic interaction between two different genes (LPA and IL6) to delve in the genetic influence and contribute to elucidate the pathophysiological mechanisms of CVDs.
Material and Methods
Studied populations
Blood samples were collected from 780 unrelated individuals (1560 chromosomes) born in Mexico with at least three generations of ancestors born in this country, who were divided into four groups. The first group (cases, n=204) and the second one (controls, n=204) were recruited from the Thrombosis, Haemostasis and Atherogenesis Medical Research Unit of the Mexican Social Security (IMSS initials in Spanish) in Mexico City.
Cases group was confirmed by individuals with myocardial infarction (58%) and stroke (42%), not taking lipid-modifying hypoglycemic drugs. Individuals with myocardial infarction (MI, average age: 54; range: 29 - 85) were defined through echocardiogram, electrocardiogram, serum enzymes and/or coronal intervention. Rather, individuals with stroke (average age: 42; range: 11 - 89) were diagnosed using computerised axial tomography scan or magnetic resonance methods.
Controls (average age: 48; range: 16 - 87) were included in the absence of any atherothrombotic events confirmed by clinical records. Classical risk factors such as diabetes type 2 (HbA1c > 6.5% and fasting glucose> 126 mg/dL), hypercholesterolemia (fasting levels of cholesterol > 200 mg/dL, LDL > 135mg/dL, HDL < 45mg/dL), hypertension (systolic blood pressure > 140 mmHg / diastolic blood pressure > 90 mmHg), Body Mass Index (BMI), use of hormones, gender, and age, were also obtained from clinical records. Physical activity, diet, family history of CVD, smoking habits, drugs and alcohol intake, were determined using questionnaires.
The third group was a Genome Control (GC) whereas the fourth group was an Ancestral Genome Control (AGC). These two control groups are powerful tools to validate the GAS in populations with complex genetic architecture as the Mexican population. These controls followed the STREGA (Strengthening the Reporting of Genetic Association Studies) statement to improve the quality of GA studies, decreasing the bias caused by statistical errors type I and II [14]. GC was confirmed by 289 Mexican Mestizos (141 men and 148 women) from the Central Region of Mexico (states of Querétaro, Guanajuato, Puebla and Mexico City). These individuals presented comparable ancestral background to the cases and controls thus GC was also a critical tool to determine the population genetic parameters (i.e., gene frequencies, Hardy-Weinberg expectation and linkage disequilibrium). AGC was included to avoid the spurious associations related to the interethnic variation. This group included 83 Mexican Native Americans from three different ethnicities: Mazahuas (n=19), Me’Phaas (n=32), and Nahuas (n=32) who ancestry is one of the most prominent in the Central Region (Table 1).
Population
Region
n
Mexican (this work)
Mexican Mestizo
Central Valley of Mexico, Mexico
289
Mazahuas
Santa Rosa Estado de Mexico, Mexico
19
Me’Phaas
Upland of Sierra de Guerrero, Mexico
32
Nahuas
Southeaster of Puebla, Mexico
32
Latin American
Mexican American
Los Angeles, California, USA
64
Puerto Ricans
Puerto Rico
104
Colombians
Medellin, Colombia
94
Peruvians
Lima Peru
85
African
Yoruba
Ibadan, Nigeria
108
Luhya
Webuye, Kenya
99
Gambian
Western Divisions, Gambia
113
Mende
Sierra Leone, Republic of Sierra Leone
85
Esan
South Nigeria, Nigeria
56
African derived
Afro-Barbadian
Barbados in Caribean, Barbados
96
Americans of African Ancestry
Southwest USA
61
European
Toscani
Tuscany, Italia
107
Finnish
Finland, Republic of Finland
99
British
England and Scotland, UK
91
Iberian
Iberia, Spain
107
European derived
Northern and Western European Ancestry
UTAH, USA
99
South Asian
Punjabi
Lahore, Pakistan
96
Bengali
Bangladesh, Bangladesh
86
Sri Lankan Tamil
London, UK
102
Indian Telugu
United Kingdom
102
Gujarati Indian
Huston, Texas, USA
102
Table 1: Populations used to descriptive analysis with a short description of each one; the comparison data was obtained from the 1000 genomes project.
Each individual signed an informed consent letter where genealogical data was included to avoid overrepresentation of alleles due to inbreeding. This GAS was conducted in agreement with the principles established by the Declaration of Helsinki and approved by the ethics committee of IMSS (R-2015-3609-20) and Center for Research and Advanced Studies (CINVESTAV, initials in Spanish) in Mexico City.
Genotyping assessment
Genomic DNA was isolated from peripheral blood leukocytes using a salting-out kit (Jena Bioscience, Jena, Germany). Four Single Nucleotide Polymorphisms (SNP) were studied (IL-6-rs1800795, LPA-rs9457951, LPA-rs10455872, and LPA-3898220) using a 5’-exonuclease TaqMan assays (Applied Biosystems, Carlsbad, CA, USA) following the manufacturer’s instructions (Figure 1). PCR cycling conditions consisted of a pre-incubation period (10 min at 95°) followed by 40 cycles (0.25 min at 92 and 1.5min at 60°) using a C1000 Touch Thermal Cycler (Bio-Rad, Richmond, CA) for data acquisition. All experiments were performed in duplicate.
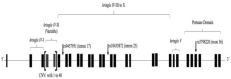
Figure 1: Lipoprotein (a) gene structure; each box depicted an exon.
Footnote. CNV: Copy Number Variable.
Statistical analysis
Population genetic analysis: Allele and genotype frequencies, as well as the Linkage Disequilibrium (LD) were done using Arlequin v.3.5 software [15]; multi-allele frequencies were estimated using direct counting. Hardy-Weinberg expectation was determined by Genetix v.4.05 software applying Weir and Cocker ham’s F statistics using 10,000 permutations [16].
To avoid spurious associations due to population stratification, the genetic differences between cases and controls were corrected with the number of subpopulations (k) previously reported by Gomez et al, using the Strat v.1.1 software [17,18].
Comparisons with other populations: To compare the genetic data obtained in the present study with African-, Asian- and European descendants, 1200 individuals from 1000 Genomes Project was employed (Table 1) [19]. The genetic differences among populations were visualised through Multidimensional Scaling (MDS) plot with SPSS v.23 software (Chicago, IL, USA) [20].
Statistical test and Models
Comparisons of statistical means between cases and controls for variables such as smoking (pack-year), physical activity (metabolic equivalent, METs; min/week), and energetic intake (kcal/day) were performed by Mann-Whitney’s test. Additionally, the statistical means differences in parametric variables such as BMI, age, cholesterol, and alcohol intake were evaluated using Student’s t-test. The genetic differences between cases and controls by categorical risk factors (i.e., hypertension, hypercholesterolemia, diabetes, and hormones uptake) were compared by chi-square test. These statistical parameters were performed with STATA v.12.0 software (Stata Corp., College Station, TX, USA) [21].
The continuous variables that exhibited differences between groups were categorized to fit logistic models, as follows: tobacco variable (non-smokers or reference group, smokers with low and high exposure according to the median control group, ≈ 2 pack per year); physical activity was categorized according to World Health Organization parameters as high (>500 METs min/week), medium (0< X =500 MET-min/week) and moderate activity; alcohol use separated into a mean control group (30.6 mg/week) and a reference group as (non-drinkers) [22].
Ultimately, multiple logistic regression models (for risk alleles) were adjusted for confounders, using the STATA v. 12.0 software. Possibly modifiers were determined by interaction analysis (data not shown). This analysis did not identify any interactions among variables.
Results and Discussion
Table 2 shows the statistical differences in the co-variables between cases and controls. Classical risk factors such as hypertension, hypercholesterolemia and tobacco exposure presented statistical differences, being consistent with previous reports [23,24]. Cigarette smoking has been related to the promotion of prothrombotic molecules secretion, reinforcing its contribution in CVD development [25]. The family history of CVD also presented statistical differences (P = 0.0001), highlighting the importance of GAS [26]. In the present study, the search of predisposition markers to delve in the genetic influence of this medical condition was done.
Variable
Cases
Controls
P Value
n = 204
n = 204
Parametric Variables (Mean± SEM)
BMI
26.6 ± 0.31
27.15 ± 0.35
0.2244
Age (years)
47.5 ± 1.01
48 ± 1.16
0.7634
Cholesterol intake (mg/day)
244.33 ± 14.48
256.56 ± 13.15
0.5329
Alcohol consumption (gr/week)
54.92 ± 11.34
30.68 ± 4.34
0.046
Non-Parametric Variables.Median [IR]
Tobacco (pack-year)
4 [1-15]
2 [0.3 - 9]
0.0074
Physical Activity (MET-min/week)
960 [0 - 3360]
960 [0 - 2160]
0.7017
Caloric Intake (kcal/day)
1722.3 [1438 - 2065]
1615.5 [1404 - 1933]
0.072
Categorical Variables
Sample size (%)
Gender (Women)
102 (50)
102 (50)
1
Stroke/MI
120 (58.8)/84 (41.2)
-
-
Hypertension
57 (27.9)
36 (17)
0.0132
Hypercholesterolemia
41 (20)
13 (6.4)
<0.0001
Diabetes
27 (13.2)
21 (10.3)
0.3566
Family history
112 (54.9)
56 (27.4)
<0.0001
Use of hormones
1 (0.49)
3 (1.4)
0.3149
Table 2: Comparisons between cases and controls using bivariate co-variable analysis.
Overall, the studied populations exhibited common tendencies both in allele as well in genotypic frequencies (Table 3). LPArs9457951 exhibited a monomorphic behaviour, however, nuance differences were found in the cases population related to the G allele (OR = 2.85, 95%CI = 0.95 – 8.60, P = 0.05). This trend was also found using an additive model where the frequency of GG+GC was higher in cases than in controls plus genomic control (OR = 3.71, 95%CI = 1.03 – 13.26, P = 0.04) suggesting a marginal association. Nevertheless, the sample size and the prominent frequency of C allele in the Hispanics limit this interpretation [12]. Meanwhile, LPA-rs10455872 exhibited similar distribution than those reported in other Latino populations [27]. On the contrary, LPA-rs3798220 presented remarkable differences in its distribution. The C allele, defined as risk allele, presented low frequencies in Brazilians (0.06), Afro-descendants, and European-derived populations (< 0.01), being more frequent in the Mexican populations [28]. Of note, the high prevalence of this allele among the Mexican Native Americans (0.42 – 0.61), suggesting a possible contribution of this ancestry to CVDs development. In concordance with this finding, the Amerindian background has been related to dyslipidaemia incidence and consequently to CVDs predisposition [29].
TABLECREATED
Table 3: Descriptive population genetic parameters in studied populations.
CVD are polygenic conditions that follow heterogeneous patterns. Thus, the study of multiple loci and different polymorphisms within each locus is one of the best approaches to explore its possible contribution in genotype-phenotype associations. In that vein, different Multi-Allelic Combinations (MACs) were built using the risk alleles addressed in prior reports. Frequency differences between cases and controls were found in two MACs. Given that the frequency in these two MACs was absent in controls, statistical limitations were presented to make any association. To solve this inconvenient, the frequencies of MACs in cases were compared with the MACs obtained in controls plus GC. Whit this approach, the MAC-1 composed of the polymorphisms rs10455872-G and rs3798220-C located on LPA gene (OR = 2.28, 95%CI = 1.02 – 5.09, P = 0.03) showed important differences. Moreover, the MAC-2 composed of LPA-rs10455872-G and IL6-rs1800795-C (OR = 4.06, 95%CI = 0.96 – 17.16, P = 0.05) presented a marginal difference also imposed by small a sample size. With this in mind, a possible contribution of these MACs with continuous variables was done. Carriers of MAC-1 presented 1.48 times more cholesterol intake than non-carriers (388 mg/day versus 217 mg/day; P = 0.03), which may contribute to 1.8 times more risk of stroke development (P = 0.02). Interestingly, this is independent of the physical activity (2407 METs versus 6805 METs; P = 0.001), underlying the importance of genetic contribution. All values were adjusted to age, diet and physical activity.
Our results support previous findings where rs10455872 and rs3798220 have been associated with CVD; thus these polymorphisms could be used as potential risk biomarkers [30,31]. Particularly, rs10455872 has been related to the circulationLp (a) levels. This phenotype-genotype relation is the consequence of the strong association between rs10455872 and the copy number of Kringletype 2 (KIV-2), which length modifies Lp (a) levels [9]. However, other SNPs located upstream of LPA gene have also been associated with CVD [12,31]. This association is also the result of the critical role of LPA in different peculiarities of CVD: thrombosis promotion, development and progression of coronary atherosclerosis, and the arterial repository of oxidised phospholipids through apo B100, among others [12,32,33]. As a consequence, variants in LPA are considered as a risk factor per se, supported by multi-ethnic studies, GWAS, and WES [34-36]. Although LPA is a causal and independent risk factor, the present study underlines the importance of gene interactions to elucidate the aetiology of these broad medical conditions. Hence, the interaction between inflammation processes, the lipid metabolism, and the endothelial dysfunction was addressed herein.
Despite no statistical differences were found in the combination between IL6-rs1800795-C and LPA-rs3798220-C, this interaction could be related to a modest gain in weight (P = 0.02). This anthropometric characteristic may increase the MI in 1.13 times, mainly in men, suggesting a possible gender-based difference. Previously, IL6-rs1800795 has been associated with CVD in different populations [37,38]. It is noteworthy that important controversies have been reported about this polymorphism, whose risk allele has not been totally defined. Prior studies in Italian, Spanish and United Kingdom groups have positively associated the C allele with CVD [39-42]. On the other hand, G allele has been reported as a risk allele by European (Austrian, Italian, and Polish) and Asiatic (Chinese) research groups [11,43-46]. These discrepancies may relate to an interethnic variation where the C allele could have been directly imported from Spain during the colony period [47]. Thus, presentday Mexican Mestizos are the carrier of this allele, which is extremely scarce in the Native Americans. In this setting, European-derived populations such as Utah residents with Northern and Western European ancestry from the Centre d’Etude du PolymorphismeHumain (CEPH, initials in French) collection (CEU) as well as Toscani in Italy (TSI) exhibit the highest frequencies supporting this hypothesis [19].
About the moderate gain of weight found in IL6-rs1800795, prior reports have shown an association of the CC genotype with high Bone Mineral Density (BMD) [48,49]. The increase in BMD is concurrent with higher weight, reinforcing our findings.
All populations were in Hardy-Weinberg Equilibrium (HWE) with a slight departure in the cases caused by homozygote excess (Fis= 0.086; P = 0.027). Although this departure was lost after the Bonferroni’s correction (P = 0.0125), it could suggest an evidence of genetic association [50,51]. This possible finding was supported by the HWE in both controls and GC, which arose from the same population than cases. Besides, the MDS plot (stress value = 0.007), where the cases exhibited a substantial genetic differentiation regarding the controls and the GC bolstered a possible association (Figure 2). However, it is likely that the small sample size may increase the homozygosity in the studied population as a result of genetic drift [52].
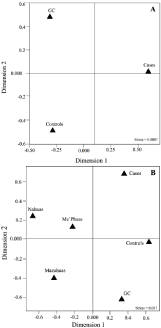
Figure 2: Multidimensional Scale Analysis showing the genetic differences
among (A) cases, controls, and GC; and (B) cases, controls, GC, and Native
Americans.
Footnote: GC: Genomic Control
The accuracy in GAS is a keystone to validate the associations. Genetic statistic parameters are powerful tools to support the phenotype-genotype associations, avoiding spurious results [53]. Thus, together with Hardy-Weinberg expectations, LD patterns were explored. A significant LD between IL6-rs1800795 and LPA-rs3798220 (P = 0.0001) was found in controls and GC but not in cases, suggesting that these differences were the result of a genetic stratification (Figure 3A). When compared our results with worldwide populations from the 1000 genomes project, different patterns were also found. African, European, South Asian and even Mexican ancestry in Los Angeles, California (MXL) populations exhibited linkage equilibrium; Peruvian in Lima, Peru (PEL) population presented LD (Figure 3B). These difference are a signature of spurious LD deeply present in admixed populations such as Mexican and Peruvian [54]. Therefore, the genetic stratification correction using Bayesian methods was also done; the phenotype-genotype relations were marginally maintained (P = 0.09) supporting the influence of genetic background in the CVD. Thus, the linkage equilibrium found in cases probably was a consequence of the small population size.
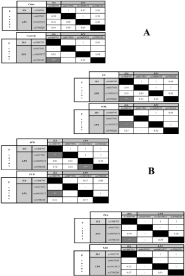
Figure 3: Linkage Disequilibriumpatterns among different populations using
the four SNPs studied (A) cases, controls, GC, and MXL; and (B) AFR, EUR,
PEL, and SAS.
Footnote. LD: Linkage Disequilibrium; D’: coefficient of LD normalized; GC:
Genomic Control; AFR: African and Afro-descendants gene pool; EUR:
European gene pool; MXL: Mexican Ancestry from Los Angeles, CA, USA;
PEL: Peruvian in Lima Peru; SAS: South Asian gene pool. P: adjusted P
value for 5% nominal level (0.000152). *: P value = 0.01; **: P value = 0.001;
***: P value = 0.0001.
Although our study presented weaknesses in the sample size, the inclusion of GC and AGC were important strengths, which were highlighted in a recent study that reinforces the present findings [12]. However, our results should be interpreted in light of these limitations.
Conclusion
The present study supports prior findings of LPA gene contribution in the development of CVD and its role as a risk biomarker. Moreover, the importance of gene interactions among different loci and the advantages of population genetics in the strengthening of the genetic associations were highlighted. To our knowledge, this is the first study that reports an interethnic variation in IL6-rs1800795.
Acknowledgment
This study was supported by Consejo Nacional de Ciencia y Tecnología (Conacyt) project FOSISS-261268 (to R.G.). We are grateful the enthusiastic participation of all populations studied herein; their collaboration made this study possible. Finally, we thank Conacyt for Scholarship 396941 granted to LirioIveth Lopez-Salazar during her Master’s in Sciences.
References
- World Health Organization. Top 10 Causes of Death. 2016.
- Yiannakouris N, Katsoulis M, Trichopoulou A, Ordovas JM, Trichopoulos D. Additive influence of genetic predisposition and conventional risk factors in the incidence of coronary heart disease: a population-based study in Greece. BMJ Open. 2014; 4: e004387.
- Auer PL, Nalls M, Meschia JF, Worrall BB, Longstreth WT, Seshadri S, et al. Rare and Coding Region Genetic Variants Associated With Risk of Ischemic Stroke. JAMA Neurol. 2015; 72: 781-788.
- O’Donnell CJ, Nabel EG. Genomics of Cardiovascular Disease. N Engl J Med. 2011; 365: 2098–2109.
- Schmidt K, Noureen A, Kronenberg F, Utermann G. Structure, function, and genetics of lipoprotein (a). J Lipid Res. 2016; 57: 1339–1359.
- Kamstrup PR. Lipoprotein(a) and ischemic heart disease-A causal association? A review. Atherosclerosis. 2010; 211: 15–23.
- Anuurad E, Boffa MB, Koschinsky ML, Berglund L. Lipoprotein(a): A Unique Risk Factor for Cardiovascular Disease. Clin Lab Med. 2006; 26: 751–772.
- Pant S, Deshmukh A, GuruMurthy GS, Pothineni NV, Watts TE, Romeo F, et al. Inflammation and Atherosclerosis? Revisited. J Cardiovasc Pharmacol Ther. 2014; 19: 170–178.
- Ferretti G, Bacchetti T, Johnston TP, Banach M, Pirro M, Sahebkar A. Lipoprotein(a): A missing culprit in the management of athero-thrombosis? J Cell Physiol. 2017.
- Wang K, Dong PS, Zhang HF, Li ZJ, Yang XM, Liu H. Role of interleukin-6 gene polymorphisms in the risk of coronary artery disease. Genet Mol Res. 2015; 14: 3177–3183.
- Greisenegger S, Endler G, Haering D, Schillinger M, Lang W, Lalouschek W, et al. The (−174) G/C polymorphism in the interleukin-6 gene is associated with the severity of acute cerebrovascular events. Thromb Res. 2003; 110: 181–186.
- Lee S-R, Prasad A, Choi Y-S, Xing C, Clopton P, Witztum JL, et al. LPA Gene, Ethnicity, and Cardiovascular Events Clinical Perspective. Circulation. 2017; 135: 251–263.
- Talmud PJ. Gene-environment interaction and its impact on coronary heart disease risk. Nutr Metab Cardiovasc Dis. 2007; 17: 148–152.
- Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, von Elm E, et al. Strengthening the reporting of genetic association studies (STREGA)-- an extension of the strengthening the reporting of observational studies in epidemiology (STROBE) statement. J Clin Epidemiol. 2009; 62: 597–608.
- Excoffier L, Laval G, SS. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinforma. 2005; 1: 47–50.
- Belkhir K, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Univ Montpellier II. 1996.
- Gómez R, Magaña J, Cisneros B, Pérez-Salazar E, Faugeron S, Véliz D, et al. Association of the estrogen receptor α gene polymorphisms with osteoporosis in the Mexican population. Clin Genet. 2007; 72: 574–581.
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000; 67: 170–181.
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491: 56–65.
- BM Corp. R. IBM SPSS Statistics for Windows. NY: IBM Corp. 2013.
- Stata Corp LP. Stata statistical software: Release 12.1. Stata Press, College Station. TX. 2012.
- Ainsworth BE, Haskell WL, Herrmann SD, Meckes N, Bassett DR, Tudor-Locke C, et al. 2011 Compendium of physical activities: A second update of codes and MET values. Med Sci Sports Exerc. 2011; 43: 1575–1581.
- Koch W, Mueller JC, Schrempf M, Wolferstetter H, Kirchhofer J, Schömig A, et al. Two Rare Variants Explain Association with Acute Myocardial Infarction in an Extended Genomic Region Including the Apolipoprotein (A) Gene. Ann Hum Genet. 2013; 77: 47–55.
- Arsenault BJ, Boekholdt SM, Dube MP, Rheaume E, Wareham NJ, Khaw KT, et al. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet. 2014; 7: 304–310.
- Barua RS, Ambrose JA, Srivastava S, DeVoe MC, Eales-Reynolds LJ. Reactive Oxygen Species Are Involved in Smoking-Induced Dysfunction of Nitric Oxide Biosynthesis and Upregulation of Endothelial Nitric Oxide Synthase: An In Vitro Demonstration in Human Coronary Artery Endothelial Cells. Circulation. 2003; 107: 2342–2347.
- Tian T, Jin G, Yu C, Lv J, Guo Y, Bian Z, et al. Family History and Stroke Risk in China: Evidence from a Large Cohort Study. J Stroke. 2017; 19: 188–195.
- Santos PC, Bueno CT, Lemos PA, Krieger JE, Pereira AC. LPA rs10455872 polymorphism is associated with coronary lesions in Brazilian patients submitted to coronary angiography. Lipids Health Dis. 2014; 13: 74.
- Thomas RK, Baker AC, DeBiasi RM, Winckler W, LaFramboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007; 39: 347–351.
- Ko A, Cantor RM, Weissglas-Volkov D, Nikkola E, Reddy PMVL, Sinsheimer JS, et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat Commun. 2014; 5: 3983.
- Cha YJ, Lee J, Choi SS. Apoptosis-mediated in vivo toxicity of hydroxylated fullerene nanoparticles in soil nematode Caenorhabditis elegans. Chemosphere. 2012; 87: 49–54.
- Mack S, Coassin S, Rueedi R, Yousri NA, Seppala I, Gieger C, et al. A genome-wide association meta-analysis on lipoprotein(a) concentrations adjusted for apolipoprotein(a) isoforms. J Lipid Res. 2017.
- Jacobson TA. Lipoprotein(a), cardiovascular disease, and contemporary management. Mayo Clin Proc. 2013; 88: 1294–1311.
- Helgadottir A, Gretarsdottir S, Thorleifsson G, Holm H, Patel RS, Gudnason T, et al. Apolipoprotein(a) genetic sequence variants associated with systemic atherosclerosis and coronary atherosclerotic burden but not with venous thromboembolism. J Am Coll Cardiol. 2012; 60: 722–729.
- Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. Cutler D, editor. PLoS Genet. 2014; 10: e1004494.
- Deo RC, Wilson JG, Xing C, Lawson K, Linda Kao WH, Reich D, et al. Single-nucleotide polymorphisms in LPA explain most of the Ancestry-specific variation in Lp(a) levels in African Americans. PLoS One. 2011; 6: e14581.
- Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, et al. Genetic Variants Associated with Lp(a) Lipoprotein Level and Coronary Disease. N Engl J Med. 2009; 361: 2518–2528.
- Liu CL, Xue ZQ, Gao SP, Chen C, Chen XH, Pan M, et al. The Relationship between Interleukin-6 Promotor Polymorphisms and Slow Coronary Flow Phenomenon. Clin Lab. 2016; 62: 947–953.
- Lima-Neto LG, Hirata RDC, Luchessi AD, Silbiger VN, Stephano MA, Sampaio MF, et al. CD14 and IL6 polymorphisms are associated with a pro-atherogenic profile in young adults with acute myocardial infarction. J Thromb Thrombolysis. 2013; 36: 332–340.
- Spoto B, Mattace-Raso F, Sijbrands E, Leonardis D, Testa A, Pisano A, et al. Association of IL-6 and a Functional Polymorphism in the IL-6 Gene with Cardiovascular Events in Patients with CKD. Clin J Am Soc Nephrol. 2015; 10: 232–240.
- Panoulas VF, Stavropoulos-Kalinoglou A, Metsios GS, Smith JP, Milionis HJ, Douglas KMJ, et al. Association of interleukin-6 (IL-6)-174G/C gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis: The role of obesity and smoking. Atherosclerosis. 2009; 204: 178–183.
- Licastro F, Chiappelli M, Caldarera CM, Tampieri C, Nanni S, Gallina M, et al. The concomitant presence of polymorphic alleles of interleukin-1beta, interleukin-6 and apolipoprotein E is associated with an increased risk of myocardial infarction in elderly men. Results from a pilot study. Mech Ageing Dev. 2004; 125: 575–579.
- Chamorro A, Revilla M, Obach V, Vargas M, Planas AM. The -174G/C polymorphism of the interleukin 6 gene is a hallmark of lacunar stroke and not other ischemic stroke phenotypes. Cerebrovasc Dis. 2005; 19: 91–95.
- Flex A, Gaetani E, Papaleo P, Straface G, Proia AS, Pecorini G, et al. Proinflammatory genetic profiles in subjects with history of ischemic stroke. Stroke. 2004; 35: 2270–2275.
- Buraczynska M, Zukowski P, Drop B, Baranowicz-Gaszczyk I, Ksiazek A. Effect of G(-174)C polymorphism in interleukin-6 gene on cardiovascular disease in type 2 diabetes patients. Cytokine. 2016; 79: 7–11.
- Antonicelli R, Olivieri F, Bonafè M, Cavallone L, Spazzafumo L, Marchegiani F, et al. The interleukin-6 -174 G>C promoter polymorphism is associated with a higher risk of death after an acute coronary syndrome in male elderly patients. Int J Cardiol. 2005; 103: 266–271.
- Jeng JR, Wang JH, Liu WS, Chen SP, Chen MYC, Wu MH, et al. Association of interleukin-6 gene G-174C polymorphism and plasma plasminogen activator inhibitor-1 level in Chinese patients with and without hypertension. Am J Hypertens. 2005; 18: 517–522.
- Sans N. Admixture studies in Latin America: From the 20th to the 21st Spec Genet Anthropol. 2000; 72: 155–177.
- Ni Y, Li H, Zhang Y, Zhang H, Pan Y, Ma J, et al. Association of IL-6 G-174C polymorphism with bone mineral density. J Bone Miner Metab. 2014; 32: 167–173.
- Magana JJ, Gomez R, Cisneros B, Casas L, Valdes-Flores M. Association of interleukin-6 gene polymorphisms with bone mineral density in Mexican women. Arch Med Res. 2008; 39: 618–624.
- Goddard KAB, Ziegler A, Wellek S. Adapting the logical basis of tests for Hardy-Weinberg Equilibrium to the real needs of association studies in human and medical genetics. Genet Epidemiol. 2009; 33: 569–580.
- Ryckman K, Williams SM. Calculation and use of the Hardy-Weinberg model in association studies. Curr Protoc Hum Genet. 2008; 1–11.
- Templeton AR. Population Genetics and Microevolutionary Theory. Hoboken, NJ, USA: John Wiley & Sons Inc. 2006.
- Hu Y, Lu Q, Liu W, Zhang Y, Li M, Zhao H. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. Zhu X, editor. PLoS Genet. 2017; 13: e1006836.
- Rousset F. Genetic Structure and Selection in Subdivided Populations. 2004; 288.