
Review Article
Austin J Vaccines & Immunother. 2015; 2(1): 1006.
Future Clinical Applications of the Potential use of Lactic Acid Bacteria as Vehicles to Deliver DNA Vaccines
Mancha-Agresti P1, Sousa CS1, Carmo FLR1, Oliveira Junior AF1, Azevedo V1 and de Azevedo MSP1*
Departamento de Biologia Geral, Universidade Federal de Minas Gerais, Brazil
*Corresponding author: Marcela Santiago Pacheco de Azevedo, Departamento de Biologia Geral, Laboratório de Genética Celular e Molecular (LGCM), Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, CP 486 CEP 31270-901, Belo Horizonte - MG, Brazil
Received: June 16, 2015; Accepted: August 27, 2015; Published: August 29, 2015
Abstract
The use of Lactic Acid Bacteria (LAB) as DNA delivery vehicles represents an interesting strategy as they are regarded as safe. Within the group of LAB, the Lactococcus lactis is deemed as a model microorganism, which is being extensively used for antigen and cytokines production and delivery to the mucosal level. Recently studies about these bacteria have focused on their usage as vehicles for the delivery of genic vaccines. Wild type or recombinant invasive L. lactis are able to trigger DNA expression by epithelial cells, both in vitro and in vivo, important for effectiveness of the vaccine. For this, invasive strains of L. lactis have been developed in order to increase the delivery efficiency of these vaccines to host cells. DNA vaccines are plasmid structures with genes that encode antigenic/therapeutic proteins or peptides capable of triggering an immune response against a wide range of diseases. This review summarizes the potential use of Lactic Acid Bacteria as vehicles to deliver DNA vaccines.
Keywords: DNA vaccine; Delivery Vectors; Lactic Acid Bacteria; Lactococcus Lactis
Introduction
The use of DNA as a strategy for vaccination has progressed very quickly since the first publication, in 1992 [1]. DNA vaccines are the third generation vaccine that contains the best-required elements of standard vaccines to be used in humans. This vaccination strategy has the ability to induce potent cellular immune responses, in addition to antibodies and the elasticity to express multiple antigens or epitopes using a single DNA vector [2]. Genetic immunization involves the transfer of a gene encoding an antigenic protein cloned in expression vectors to a eukaryotic cell from the host, leading to the induction of an immune response against the expressed antigen [3]. Therefore theses transfected mammalian cells are able to express in situ the antigen (for vaccines) or the therapeutic protein (for gene therapy applications) [4]. Furthermore, they do not have the inconvenient of classical vaccines: they are safe, inexpensive, easy to produce, heat stable and amenable to genetic manipulation [3]. The DNA vaccine is composed of a plasmid backbone that contains a bacterial origin of replication needed for the vector’s maintenance and propagation inside the bacteria, as well as a resistance marker, necessary to permit a selective growth of the bacteria that carries the plasmid; immunostimulatory sequences (ISS), for example, the “CpG motifs” (cytosine-phosphate-guanineunmethylated). They are responsible for increasing the magnitude of the immune response as they can enhance T lymphocyte recruitment or expansion [5–8]. Moreover, these ISS sequences can interact with Toll-like receptors (TLR), such as TLR9, and add adjuvant activity [9]. Another component of DNA vaccines is the transcriptional unit, necessary for eukaryotic expression, which harbors a promoter/enhancer region, introns with functional splicing donor and acceptor sites, as well as the ORF (open reading frame) encoding the antigenic protein of interest, and the polyadenylation sequence (poly A), signal required for efficient and correct transcription termination of the ORF and transfer of the stable mRNA from the nucleus to the cytoplasm [3,10]. The polyA sequence usually is derived from the bovine growth hormone, SV40, or rabbit β-globin gene [8]. The ORF encoding the protein of interest contains a Kozak translation initiation sequence (ACCATGG) harboring an initiation codon (ATG) for appropriate translation [11–13]. The insert also contains a termination codon (TAA, TGA, or TAG) that signals a termination of translation. Major structures of DNA vaccines are illustrated in (Figure 1).
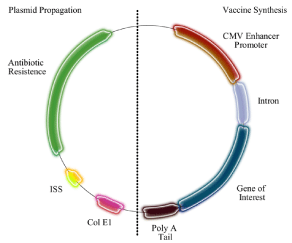
Figure 1: Genetic element of a plasmid DNA vaccine. The vaccine
synthesis (eukaryotic expression region) harbors the enhancer/promoter,
introns, transgene of interest and a transcriptional terminators (poly A), which
together lead the protein synthesis. The Plasmid propagation in the microbial
host comprises a prokaryotic origin of replication (ColE1) and a selectable
marker and the immunostimulatory sequences (ISS).
The promoter is the element used to drive the expression of the transgene/antigen in eukaryotic cells. The most broadly used promoter in traditional DNA formulations is the human cytomegalovirus (hCMV), which induces strong and constitutive expression of a protein in a variety of cell types [14,15]. An alternative to these viral promoters is the use of both human polyubiquitin C (UbC) and the elongation factor 1 α (EF1 α) promoters, which has been shown to persistent gene expression in mouse lung cells, leading to a fourfold increased protein expression level of an antigenic protein when compared to viral promoters [16]. Another element found in DNA vaccines localized after the promoter sequence and before the ORF from the gene of interests are the introns. These genetic elements were reported to increase promoter activity [6] and to avoid antigen expression by the prokaryotic machinery from bacteria, turning possible the Heterologous expression of the protein only by the eukaryotic system [7].
Plasmid backbones commercially available approved for gene therapy and vaccination include pVax1, pVAC, pDNAVACultra and pcDNA and others. They contain an Escherichia coli origin of replication such as pUC or pBR322, which allow plasmids to replicate in bacterial cell generating many copies of the plasmid in a short period of time [17]. The plasmids used in gene therapy and vaccination preferentially have TN903 gene, coding for amino glycoside enzyme that confers resistance to kanamycin, an antibiotic that is not widely used in humans preventing the risk of allergic responses when compared to others antibiotics [18]. DNA vaccines have a broad range of features that offer them many advantages over other vaccination platforms. The principal advantage concerning the DNA vaccine refers to the fact that they are easy to handle and rapid to construct. This is a fascinating attribute when considering an emerging pandemic threat [4]. In addition to this property DNA vaccines are (i) safe as plasmids do not replicate in human cells do not have the potential to integrate into the human cellular DNA (ii) No adverse effects have been reported neither tolerance to the antigen nor autoimmunity [19]; (iii) They have been shown to stimulate immunity through MHC I-mediated antigen presentation triggering both proliferation and activation of T and B cells antigen-specific; (iv) Vaccine manufacturing is simple and low cost as it requires only cloning techniques in order to clone the protein of interest; (v) they are stable at room temperature, easy to store and transport, presents thermal stability and long life time [20,21]. To sum up, DNA vaccines represents an attractive tool due to its property to induce all three points of adaptive immunity: antibodies, helper T cells (TH) and cytotoxic T lymphocytes (CTLs), as well as being capable of stimulating innate immune responses [22].
Immunological Features of DNA Vaccine
Routes of administration
The therapeutic delivery of nucleic acid has recently been recognized as a promising tool for the treatment of several infectious and genetic diseases. In order to exert their protective effects, DNA vaccines needs a suitable delivery technology to produce a desired immune response.
Intramuscular injection is the most broadly used method to administrate DNA vaccine. Nevertheless, it has been shown that this method is inefficient to induce immune response in large animals as well as in humans for some reasons: the plasmid DNA administered by this manner is inefficiently expressed, poorly distributed, and rapidly degraded [23]. The low immunogenicity of DNA vaccines observed in these studies with humans and primates have indicated scientist to focus on other methods of administration where antigenpresenting cells (APCs) would be transfected in a higher efficiency and stimulating stronger immune response [24,25]. Considering the disadvantage of intramuscular injection, other possible routes of administration have been studied, such as intradermal, subcutaneous, intraperitoneal, sublingual, intrarectal, ocular, and application of the DNA vaccines to mucosal surfaces (vaginal, nasal and oral) [26]. The pathway of administration has been shown to influence the nature and the power of immune responses. Mucosal immune responses are most efficiently induced by administration of vaccines to the mucosa, where as systemic immunization strategies rarely induce long lasting or optimal mucosal immunity and are therefore less effective against infection at the mucosal surfaces.
Course of plasmid inside the cell and development of immunogenicity
Once the DNA vaccine is administrated, the DNA plasmid will transfect different types of cells, for instance myocytes, keratinocytes, resident Antigen Presenting Cells (APCs), such as dendritic cells (DCs) and macrophages [4,27]. Inside the cells, the plasmids will reach the nucleus surviving to the attack of endonuclease, using a microtubule net [28]. Inside the nucleus, the plasmid DNA has contact to the transcription machinery, which allows the transcription of the gene of interest [4,26]. The host cell offers the necessary posttranslational modifications mimicking a real infection. The Bacterial mediated transfer for plasmid DNA is illustrated in Figure 2.
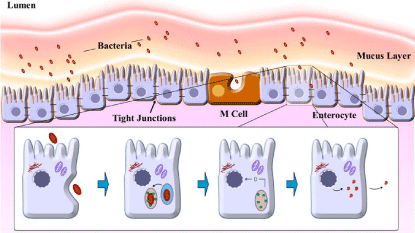
Figure 2: Schematic representation of bacterial mediated transfer
of plasmid DNA into mammalian cells. First, the entrance of bacteria
in mammalian cells. Second, the phagolysosome engulfs the bacteria to
cleaves it. Third, the DNA plasmid escapes from the vesicle and reaches
the nucleus of the mammalian cells. Fourth, in the nucleus the transgene
is transcribed and the protein synthesis is realized by host cell. Finally, the
protein is exposed the immune system.
This feature is one of the biggest advantages of the genetic immunization [4,27]. The produced proteins are then presented to the surface of the cells becoming a target to the immune system. Antigenic proteins can be secreted as well generating both humoral and cellular immune responses. The direct transfection of the DNA plasmid to the APCs cells has a critical role in DNA vaccine immunogenicity. DCs are probably the most important APCs associated with the capture and processing of antigens via receptor- mediated endocytosis as it presents antigens trough MHC class I or II directly to T naïve cells, leading to TCD4+ and TCD8+ lymphocytes activation. The stimulation of this immunological repertoire raises both humoral and cellular protection against the antigen inside the host [29,30].
Clinical advance in DNA vaccine platform
FDA still did not approve the use of DNA vaccines in humans. Phase I clinical studies have been reported for the prevention and/ or treatment of HIV, malaria, hepatitis B, SARS and many other infectious agents [31]. In order to strongly trigger the innate immune mechanisms and to go guarantee an optimal expression of the antigen, it is essential to efficiently deliver the DNA into the cells ensuring their transfer to the nuclei [32]. Actually, the inefficient uptake/transfer of the plasmids into mammalian cells is one of the principal hurdles in DNA vaccinology [21]. The lack of reproducibility of many results obtained in mice after the applications in larger animals, as well as the failure of DNA vaccines to induce potent immune responses in humans have not yet been elucidated [33]. For this reason, novel strategies to improve transfection efficiency are emerging. Among them, can be highlighted the delivery methods employed to introduce the DNA vaccine in the organism [33]. These methods include physical and chemical approaches or the biological strategies, such as the use of viral and bacterial vectors. The physical methods of delivering include: gene gun, tattooing, ultrasound (US), electroporation (EP), laser and dermal patches. The chemical vectors most studied are lipid and polymer complexes. The principal advantages of these vectors are that they can tightly compact and protect the DNA and be recognized by specific cell-surface receptors expressed in DCs or macrophages. Moreover, they can disrupt the endosomal membrane to deliver DNA plasmids allowing their transfer to the nucleus when the pH of the medium is reduced to below to six [33]. Even though physical and chemical vectors have shown to be interesting tools to delivery DNA, other methods are being explored as well, such as viruses and bacteria. Regarding the use of viruses as delivery vehicles, a severe adverse effect has occurred during in a gene therapy trial raising serious safety concerns. Attenuated viruses have been studied for gene therapy/antigen delivery, however it was shown that their genome could integrate the host cellular chromatin (oncoretrovirus and lentivirus), which may cause genetic diseases or favor the development of cancerous cells [34].
Regarding the use of biological vehicles, bacterial vectors have been shown to be an excellent choice, as they can transfer the genetic material into mammalian cells such as viruses and does not present the same problems associated with their use. Both attenuated bacterial pathogens or live recombinant bacteria are considered good models for DNA delivery due to their ability to protect the DNA vaccine from enzymatic digestion, to stimulate the immune system as they can target inductive sites of the body generating effective adaptive immunity [35].
Bacterial as delivery vectors for DNA vaccines
The innate tropism of some pathogenic bacterial strains have for specific tissues of the host directed the attention of researchers to study them as a vehicle to deliver DNA vaccine, as this characteristic is indispensable for the elicitation of immune response. Several advantages can be highlighted: they can keep the plasmid in a high copy number, they are easy to manufacture, they are less laborious and has low cost as there is no need to amplify and purify the plasmid before [7,36], large-size plasmid are able to be housed inside the bacteria, permitting the insertion of multiple genes of interest, and the bacterial cell can protect the DNA against endonuclease degradation [37,38].
Additionally to these features of bacterial vectors is the possibility to use them for oral administration, important feature to stimulate both mucosal and systemic immune responses [39]. Considering the increased numbers of vaccines administered all over the word, the fact that they can be inoculated without the use of a needle turns them a cheap and safe method [40].
DNA delivery from bacterial to mucosal surfaces: immunological features
Bacteria carrying a DNA vaccine are able to cross the intestinal barrier, mainly via M cell (specialized epithelial cells named Micro fold cells) overlying Peyer’s patches (PPs). The PPs are isolated lymphoid follicles in draining gut mesenteric lymph nodes, considered more accessible to antigens and bacteria present in the luminal compartment. Another manner by which bacteria may have access to the body is through immature dendritic cells (DC) that reside in PPs. They are capable to open tight junctions between epithelial cells, extend their dendrites outside the epithelium and directly sample bacteria, thereby monitoring the contents of the intestinal lumen [41]. Moreover, bacterial vectors are able to enter inside the host body by invading intestinal epithelial cells (IECs) lining mucosal surfaces through the expression of some proteins named invasins. This characteristic refers to the capacity of attenuated pathogenic vectors to deliver DNA vaccines as they naturally produce invasins.
Regarding the vectors based on attenuated pathogenic species, once inside the cells they have the ability to escape from the phagolysosome vesicles by the secretion of a variety of phospholipases and pore-forming cytolysins and enter the cytoplasm of the host cells [36,37]. The plasmids can then reach the nucleus through the microtubules net; once in the nucleus using the host cell’s transcription machinery the protein of interest carried by the plasmid can be encoded, translated, and secreted afterwards [36,42]. The antigenic proteins may be secreted outside the cell or be presented on the surface of epithelial cell or DCs. The Major Histocompatibility Complex (MHC) class-II, from APCs, presents the exogenous proteins, turning naïve T cells activated into T helper cells (CD4+ T-cells). Furthermore, the exogenous protein may also be processed into small peptides, which are then presented on the surface of MHC class-I molecules to cytotoxic T-cells (CD8+ T-cells), stimulating them [43].
Other important components of immunity are the pattern recognition receptors (Toll-like and Nod-like receptors) expressed by IECs, B-lymphocytes and DCs that are located in the sub epithelial lamina propria. These receptors are able to recognize some bacterial components known as microbe-associated molecular patterns (MAMPs), triggering intracellular signaling pathways that lead to cytokine secretion and immune cell activation [44,45]. The production of a merged immune response encompassing the induction of humoral and cell mediated immunity (CMI) effectors like CD8+ and CD4+ T cells after DNA vaccination using bacterial vectors is a well established event known as cross-priming [46,47]. The bacterial recognition by the immune system modulates the innate immune response, therefore, supporting a vigorous and lasting adaptive response [37].
Another important characteristic is the communication between activated immune cells localized in the mucosal surface with the systemic blood where they can travel around the body via the lymph [48]. The production and secretion of antigen-specific secretory immunoglobulin A (SIgA) responses by plasmocytes is another important advantage to be considered when using bacteria as mucosal delivery vehicles for DNA vaccines.
Commensal bacteria can interact with IECs and deliver tolerogenic signals that are transmitted to the underlying cells of the immune system [49]; consequently, they are not ignored by the intestinal immune system. The gastrointestinal mucosa offers immunological tolerance against nonpathogenic bacteria and antigens; this phenomenon avoids reactions against proteins and commensal bacteria. Thus, mucosal tolerance protects the mucosa from detrimental inflammatory immune responses [50]. Actually, allergic disease development and cancer, especially colon cancer, has been associated with alterations in the intestinal micro biota [51].
Bacteria as a delivery vector for gene transfer
Salmonella typhi, Listeria monocytogenes, Shigella Flexner, Yesinia enterocolitica and Escherichia coli are the principal enteropathogenic species most widely used as bacterial delivery systems into mammalian cells [36] due to their natural tropism for DCs and macrophages in the lymphoid tissue of the intestinal mucosal surface [7]. There are several reports showing the use of these strains as bacterial vectors. For example: Salmonella typhimurium is probably the most broadly used bacteria for antigen delivery applications, such as encoding duck enteritis virus UL24 [52], encoding HIV gp140 [53], among others; Yesinia enterocolitica encoding Brucella abortus antigens [54]; Listeria monocytogenes as a gene delivery vector for targeting cancer cells [55] as it could effectively target tumors expressing surface bound antigens (Her2/neu), intracellular antigens (HPV-16 E7) or secreted antigens (PSA) [56].
The use of human enteric bacterial strains, as a bacterial carrier, is being considered an advantage because of their capacity to infect human colonic mucosa after oral administration. However, for this proposal enteropathogenic species needs to be attenuated or inactivated. Nonetheless, attenuated strains has a restrict use as they present the risk to revert to the virulent phenotype compromising its safety, as reported for the oral polio vaccine (e.g., Sabin 3) [57]. Therefore, World and Health Organization (WHO) does not recommend their use in children and immunocompromised individuals. Thus, to counteract this severe problem, it has been investigated the use of non-pathogenic bacteria, such as LAB as vectors for genetic immunization [58].
Lactic acid bacteria
Lactic acid bacteria (LAB) are distributed in diverse ecological niches; for example in plants, fermented foods, as well as in the gastrointestinal tract of many animals, including humans, where some species can live as commensal microorganisms [59]. LAB comprises mainly species of Lactobacillus, Lactococcus, Leuconostoc Oenococcus, Pediococcus and Streptococcus [60,61]. They are widely used in the industry for the production and the preservation of foods due to their ability to acidify the medium (pH 3.5 to 4.5). Therefore, LAB are ingested diary by humans in different types of foods such as cheese, wine, yogurt, fermented milks, pickles, kefir, butter, among others. It have been reported the importance of enteric lactic bacteria during their life into the human gastrointestinal tract, as they have an important impact on host metabolism, participating in microbialmammalian co-metabolism [62]. Therefore, FDA has granted some strains of LAB group as ‘‘Generally Recognized as Safe’’ (GRAS) for human consumption [63]. Another important characteristics of LAB are their capacity to restore the normal intestinal flora, eliminate intestinal pathogens, reinforce the intestinal barrier capacity to foreign antigens, stimulate nonspecific immunity such as phagocytosis, stimulation of humoral immunity and production of anti-inflammatory products [64,65]. Due to all these positive effects, some strains are considered as probiotics because when consumed at adequate levels they can positively influence the human health. Thus, scientific is extensively exploring these probiotics as an alternative treatment for some diseases, and to serve as a tool for genetic immunization [66].,
L. lactis is the best-characterized member within the LAB group being considered the model organism. They are facultative anaerobic, catalase negative, and do not form endospores. Lactococcus lactis ssp. lactis was originally found as a milk-souring isolate, but it is also associated with plants. Lactococcus lactis ssp. cremoris is used as a starter culture for the manufacture of Cheddar cheese, in which it contributes with highly prized flavor [60]. L. lactis does not colonize the digestive tract of men and animals. Besides to this economic significance of L. lactis, other important features have been described: (i) it has a completely sequenced genome [67]; (ii) they are “GRAS”, (iii) it is genetically easy to manipulate; and (iv) many genetic tools have already been developed for this species [68], and (v) does not contain Lip polysaccharide (LPS) avoiding endotoxin shock after being administered to humans [69].
Lactococcus lactis: heterologous protein delivery
Besides its traditional and safe use in the food industry, L. lactis is presenting to be a very an interesting tool to be used as a “cell factory” for the high-level protein production. This new role assigned for L. lactis is due to the fact that this bacterium does not produce endotoxins or other toxic metabolic products [70]; few proteins are secreted in L. lactis; Usp45 (Unknown Secreted Protein of 45 kDa) is the only one on e secreted in quantities large enough to be detected in Coomassie-stained protein gels [71]. This feature is very important, as it simplifies purification step after bacterial growth in fomenters [72].
Transformation protocols, cloning or screening-vectors, mutagenesis systems, protein expression and targeting-systems are examples of the availability of genetic tools that have been developed for L. lactis. These tools have been used to engineer L. lactis to produce intra- or extra-cellular recombinant proteins of viral, bacterial or eukaryotic origins [72]. These tools also allow the expression of these proteins in a controlled manner. To this end, various vectors containing constitutive or inducible promoters PlacA [73]; PnisA [74]; PT7 [75]; P170 [76]; P59 [77]; PxylT [78]; Pzn [79] have been developed and represent the basis of all expression systems in L. lactis and other LAB [80].
Nisin Controlled Gene Expression (NICE) is the most used amongst all expression systems developed for use in LAB. It offers significant potential for regulated gene expression. NisR and NisK genes encode a two-component system (NisRK), which controls the expression of the nisA gene via signal transduction. NisK functions as a membrane sensor that detects extracellular nisin. The signal is transferred to NisR through a phosphorylation process turning this gene capable of activating gene transcription, which is controlled by the PnisA promoter [74].
Therefore, in the presence of Nisin the promoter can induce the transcription of the molecule of interest [81] (Figure 3). NisK and NisR genes were isolated from the nisin gene cluster and inserted into the chromosome of L. lactis subsp. cremoris MG1363 (nisinnegative), creating NZ9000 strain (nisin positive) [82,83]. Depending on the presence or absence of the corresponding targeting signals, the protein is expressed into the cytoplasm, anchored to the cell wall or secreted to the extracellular medium. Many exogenous proteins have been expressed in L. lactis through this system [74,84,85]. NICE system has been tested in other LAB, such as Leuconostoc lactis, Lactobacillus Helveticas, Streptococcus sp., Bacillus sp., Enterococcus sp. [86] and Lactobacillus Plant arum [87], proving its versatility.

Figure 1: Schematic representation of the NICE system development
in L. Lactic. The expression of genes in response to nisin stimulus involves
a membrane-located sensor protein (NisK) and a cytoplasmic response
regulator (NisR) that controls transcriptional activation from the misA
promoter (PnisA). The auto-induction mechanism of nisin is used as a
controlled heterologous gene expression system in L. Lactic.
Another expression and targeting system that allows different cellular locations of the gene of interest for use in L. lactis is the xylose-inducible expression system (XIES), which is based on the use of a xylose inducible lactococcal promoter, PxylT from L. lactis NCDO2118 strain. In the presence of sugars (glucose, fructose and/ or mannose) PxylT is tightly repressed. However, in the presence of xylose, PxylT is transcription ally activated [88,89]. Staphylococcus aurous nuclease genes (nuc) fused or not to the lactococci Usp45 signal peptide was adopted to test the capacity of this system to express the cytoplasm or secreted form of nuc protein. Xylose-inducible nuc expression was found to be tightly controlled resulting in high-level, long-term protein production, and correct targeting either to the cytoplasm or to the extracellular medium. This expression system is versatile and can be easily switched on or off by adding either xylose or glucose, respectively [78]. XIES system has been employed in the biotechnology field for production of different heterologous protein, for example: 65-kDa heat shock proteins (HSPs) of Mycobacterium leprae [90]; S-layer protein (SlpA) of Lactobacillus brevis [91], among others.
Lactococcus lactis as live mucosal delivery vectors for vaccine
Regarding L. lactis as a vehicle to deliver DNA vaccines, many interesting features can be highlighted: (1) it was proved in different laboratories all over the world that they can carry recombinant plasmids and express antigens and therapeutic molecules at different cellular localizations [73,92]; (2) it was successfully demonstrated that L. lactis can deliver DNA into eukaryotic cells and in vivo to mice IECs [93–95]; (3) they can induce both systemic and mucosal immunity when administrated at mucosa surfaces [96,97]; (4) they can resist to the acid environment of the stomach, being able to survive into the gastrointestinal tract, ensuring recombinant protein or plasmid delivery [20]. Because L. lactis is not very immunogenic, it can be orally administrated multiple times [98], regarding its extraordinary safety profile [99]. Furthermore, L. lactis has the ability to stimulate the phagocytic system of the host [100]. All this characteristics turns it a good option for being used in immunization programs [101]. In accordance with the benefits of L. lactis, several researchers have developed different recombinant strains to be used for genetic immunization. Table 1 summarizes some studies that were performed in different laboratories around the world corroborating the ability of L. lactis in inducing long-lasting immune responses.
Antigens Expressed by L. lactis
Immune Response
Observed
Application
Reference
EDIII antigen from dengue virus type 2
Neutralization of the virus in vitro
Dengue virus
control strategy
Sim et al., 2008 (107)
Envelope protein of the human immunodeficiency virus 1 (HIV-1)
High levels of IgG and IgA antibodies against the antigen observed
HIV vaccine
Xin et al., 2003 (108)
Antigenic protein of
Proteus mirabilis (HBPM)
Protection to the animals against challenge with P.mirabilis virulent strain
Control of urinary
tract infections
Scavone et al., 2007 (109)
PspA antigen derived
from Streptococcus pneumoniae
Better protected
against challenge with the virulent strain
Vaccine against
pneumonia
Hanniffy et al., 2007 (110)
Antigen lcrV from Yersinia
pseudotuberculosis
Humoral and cellular immune responses, conferring protection against challenge
Vaccine against Far
East scarlet-like fever
Daniel et al., 2009 (111)
EspB antigen from the type III secretion
system (T3SS) of E. coli serotype O157:H7
Significant levels of
specific serum Ig and
faecal IgA
New strategie to fight against enterohemorrhagic E. coli
Ahmed et al., 2012 (112)
Leishmania antigen LACK and the proinflammatory cytokine IL-12
Protection against
challenge
Vaccine to combat
Leishmaniosis
Hugentobler et al.,
2012 (113)
Mature murine IFNgamma
--
Adjuvant tool
Bermúdez-Humarán
et al., 2008 (114)
Antigen envolved with LPS transport (wzm) from Vibrio cholerae O1 strain
Increase of systemic and mucosal immunity
Vaccine against cholera
Zamri et al., 2012 (115)
Catalase-producing L. lactis
Inhibition of chemically induced
colon cancer
Cancer therapy
De Moreno de
LeBlanc et al., 2008 (116)
IL-10
Anti-inflammatory effects controlling
intestinal inflammation
Treatment of allergy
and inflammatory
bowel diseases (IBD)
Cortes-Perez et al., 2007 (117); Braat et al. 2006(118) ; Marinho et al., 2010 (119)
Table 1: Antigens expressed by L. lactis, inducing long-lasting immune responses.
L. lactis expressing invasions for DNA delivering
Studies demonstrated both in vitro [98] and in vivo [93] that wildtype (wt) L. lactis could be used as a vector for genetic immunization. However, the percentage of gene transferred observed was low, as well as a low and transitory Th1-type immune response after immunization trials [93]. For this reason, with the aim to increase the capacity of lactococci to deliver DNA, some strains of L. lactis expressing invasins have been developed. The first engineered L. lactis to express invasins was reported in 2005 by Guimarães and co-workers; InlA gene from L. monocytogenes was cloned and expressed under transcriptional control of the native promoter. This work showed that recombinant lactococci could efficiently express the cell wall anchored form of InlA, and the invasion rates of LL-InlA+ strain in Caco-2 cells was approximately 100-fold higher than the wt lactococci. Moreover, after oral inoculation in guinea pigs, this recombinant strain was capable to invade intestinal cells [94]. However, the use of LL-InlA+ strain showed to be inconvenient because InlA cannot bind to its receptor in mice, murine E-cadherin, thus limiting the in vivo studies as the effect of LL-InlA+ strain was only possible to be explored in guinea pigs or transgenic mice, which may be laborious and/or expensive [102]. To improve this strategy another invasive strain of L. lactis was constructed by cloning the gene encoding fibronectin-binding protein A (FnBPA), from Staphylococcus aureus (LL-FnBPA+) [103]. FnBPA production at the surface of L. lactis increases invasiveness of the cells 1000 fold and increased plasmid transfer 30-fold in vitro (Innocent in et al., 2009). L. lactis InlA+ and L. lactis FnBPA+, showed comparable internalization rates in Caco-2 cells [93]. Therefore, another recombinant invasive lactococci was developed producing a mutated form of Internal in A (mInlA), appropriate to be used in a murine model [90].
In order to evaluate recombinant L. lactis expressing invasins a new vector have been developed resulted from the co-integration of two replicons: one from E. coli and the other from L. lactis, named pValac (Vaccination using Lactic acid bacteria). The pValac is formed by the fusion of (i) cytomegalovirus promoter (CMV), that allows the expression of the antigen of interest in eukaryotic cells, (ii) polyadenylation sequences from the bovine Growth Hormone (BGH), essential to stabilize the RNA transcript, (iii) origins of replication that allow its propagation in both E. coli and L. lactis hosts, and (iv) a chloramphenicol resistance gene for selection of strains harboring the plasmid. The functionality of pValac was observed after transfecting plasmids harboring the gfp ORF into Pk15 cells. The vector demonstrated to be functional as PK15 cells were able to express GFP protein. Moreover, invasiveness assays of L. lactis inlA+ carrying pValac: gfp into Caco-2 cells showed that this strain could deliver the vector to epithelial cells, in vitro [104]. This assay demonstrated that L. lactis expressing invasins and harboring functional plasmids can serve as tools for genetic immunization [93].
Besides the effort to construct invasive L. lactis strains and plasmids to be used for genetic immunization, other works have attempted to test L. lactis in the vaccination field.
De Azevedo and co-workers used L. lactis expressing a mutated form of L. monocytogenes Internal in A (LL-mInlA) carrying pValac: BLG. BLG is the bovine β lacto globulin, a major cow’s milk allergen. They were able to show the production of mInlA enhanced invasivity and allowed plasmid transfer in a higher efficiency in in vitro experiments. Besides than, were done in vivo experiments showing slightly increased plasmid transfer after oral administration [90].
An elegant work has been done with recombinants L. lactis FnBPA, L. lactis mInlA and wt strains (L. lactis non invasive), all of them carrying pValac: BLG. It was showed that the intranasal immunization of L. lactis non invasive strain carrying pValac: BLG elicited a TH1 immune response. However, when immune response elicited by L. lactis FnBPA and L. lactis mInlA were evaluated, both strains carrying pValac: BLG, it was observed the secretion of IL-4 and IL-5 in medium of BLG reactivated splenocytes, after both oral and intranasal administration. It was concluded that non invasive lactococci elicits a Th1 immune profile while the immunization with the recombinant invasive strains elicited a Th2 immune response [105].
Another work using L. lactis as DNA delivery vehicle involved the construction and evaluation of a DNA vaccine against Tuberculoses. In this work BALB/C mice were orally administered with L. lactis expressing FnBPA carrying pValac coding for the 6-kDa early secreted antigenic target (ESAT-6) gene of Mycobacterium tuberculosis. This report showed significant increase of interferon gamma (IFN-γ) production in spleen cells, as well as a significant increase of specific secretory immunoglobulin A (SIgA) production in colon tissue and fecal extracts [20].
The administration of L. lactis FnBPA, has been used for the administration of other therapeutic molecule, such as IL10, for the treatment of Inflammatory Bowel Disease (IBD), an inflammatory condition of the TGI being presented in two forms: Ulcerative colitis and Crohn’s disease. To this end, the therapeutic effect of L. lactis expressing FnBPA carrying pValac coding for Interleukin-10 from Mus musculus was evaluated using a model of acute trinitrobenzenesulfonic acid (TNBS)-induced colitis in mouse. It was observed a decrease in the severity of the inflammation with lower macroscopic and microscopic inflammatory scores in the large intestine, decrease of microbial translocation to the liver and less body weight loss. Furthermore, they were able to shown a decrease in antiinflammatory cytokine, IL-17. This work suggested that recombinant L .lactis expressing FnBPA invasins (pValac:il-10) is a suitable option to maintain an anti-inflammatory status in the GIT, especially for chronic Crohn’s disease patients [106].
Conclusion
Food-grade bacteria, such as LAB, have recently been proposed as a vehicle to express recombinant antigens and therapeutic molecules, as well as to deliver DNA vaccines. One of the LAB models, L. lactis, has been shown to act as DNA delivery vehicles and to deliver many different proteins with biomedical and biotechnological interest at the mucosal level. In spite of the necessity of other studies, the approach of using wild type or recombinant L. lactis as a tool to deliver therapeutic plasmids can be considered a promising future. An important point regarding the use of L. lactis as a DNA delivery vector is the strategy to use L. lactis expressing invasins. As recombinant invasive strains improved the delivery of DNA, especially in vitro, scientists explored new horizons testing them for many applications, for instance in the vaccination area. To date, there are no DNA vaccines available to be administered in humans. The only licensed DNA vaccine is for veterinary purposes. Being L. lactis safe for use in humans and capable to efficiently deliver DNA vaccines, it Fs use as immunization vehicles is an excellent choice for clinical applications in near future. Especially because this bacterium is easy to handle, GRAS and has a large number of genetic tools already developed.
Recombinant strains, as well as new vectors have attracted researcher’s attention and a great number of studies are in progress with the aim to develop systems using LAB to deliver DNA vaccine.
References
- Wolff JA, Dowty ME, Jiao S, Repetto G, Berg RK, Ludtke JJ, et al. Expression of naked plasmids by cultured myotubes and entry of plasmids into T tubules and caveolae of mammalian skeletal muscle. J Cell Sci. 1992; 103: 1249–1259.
- Wells J. Mucosal vaccination and therapy with genetically modified lactic acid bacteria. Annu Rev Food Sci Technol. 2011; 2: 423–445.
- Azevedo V, Levitus G, Miyoshi A, Cândido AL, Goes AM, Oliveira SC. Main features of DNA-based immunization vectors. Braz J Med Biol Res Rev Bras Pesqui Médicas E Biológicas Soc Bras Biofísica Al. 1999; 32: 147–153.
- Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev. 2011; 239 :62–84.
- Glenting J, Wessels S. Ensuring safety of DNA vaccines. Microb Cell Factories. 2005; 4: 26.
- Kano FS, Vidotto O, Vidotto MC. DNA vaccines: general concerns and its applications in human and veterinary medicine. Semina Ciênc Agrár. 2007; 28: 709.
- Becker PD, Noerder M, Guzmán CA. Genetic immunization: bacteria as DNA vaccine delivery vehicles. Hum Vaccin. 2008; 4: 189–202.
- Williams JA, Carnes AE, Hodgson CP. Plasmid DNA vaccine vector design: Impact on efficacy, safety and upstream production. Biotechnol Adv. 2009; 27 :353–370.
- Tudor D, Dubuquoy C, Gaboriau V, Lefèvre F, Charley B, Riffault S. TLR9 pathway is involved in adjuvant effects of plasmid DNA-based vaccines. Vaccine. 2005; 23: 1258–1264.
- Kowalczyk DW, Ertl HCJ. Immune responses to DNA vaccines. Cell Mol Life Sci CMLS. 1999; 55: 751–770.
- Kozak M. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987; 15: 8125–8148.
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991; 115: 887–903.
- Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990; 87: 8301–8305.
- Thomsen DR, Stenberg RM, Goins WF, Stinski MF. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci U S A. 1984; 81: 659–663.
- Schmidt EV, Christoph G, Zeller R, Leder P. The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol Cell Biol. 1990; 10: 4406–4411.
- Gill DR, Smyth SE, Goddard CA, Pringle IA, Higgins CF, Colledge WH, et al. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1alpha promoter. Gene Ther. 2001; 8: 1539–1546.
- Ingolotti M, Kawalekar O, Shedlock DJ, Muthumani K, Weiner DB. DNA vaccines for targeting bacterial infections. Expert Rev Vaccines. 2010; 9: 747–763.
- Lara AR, Ramírez OT, Wunderlich M. Plasmid DNA production for therapeutic applications. Methods Mol Biol Clifton NJ. 2012; 824: 271–303.
- Liu MA, Ulmer JB. Human clinical trials of plasmid DNA vaccines. Adv Genet. 2005; 55: 25–40.
- Pereira VB, Zurita-Turk M, Saraiva TDL, De Castro CP, Souza BM, Mancha Agresti P, et al. DNA Vaccines Approach: From Concepts to Applications. World J Vaccines. 2014; 04: 50–71.
- Grunwald T, Ulbert S. Improvement of DNA vaccination by adjuvants and sophisticated delivery devices: vaccine-platforms for the battle against infectious diseases. Clin Exp Vaccine Res. 2015; 4: 1–10.
- Li L, Saade F, Petrovsky N. The future of human DNA vaccines. J Biotechnol. 2012; 162: 171–182.
- Wang S, Zhang C, Zhang L, Li J, Huang Z, Lu S. The relative immunogenicity of DNA vaccines delivered by the intramuscular needle injection, electroporation and gene gun methods. Vaccine. 2008; 26: 2100–2110.
- Condon C, Watkins SC, Celluzzi CM, Thompson K, Falo LD. DNA-based immunization by in vivo transfection of dendritic cells. Nat Med. 1996; 2: 1122–1128.
- Porgador A, Irvine KR, Iwasaki A, Barber BH, Restifo NP, Germain RN. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J Exp Med. 1998; 188: 1075–1082.
- Faurez F, Dory D, Le Moigne V, Gravier R, Jestin A. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine. 2010; 28: 3888–3895.
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008; 9: 776–788.
- Vaughan EE, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol Ther J Am Soc Gene Ther. 2006; 13: 422–428.
- Payette PJ, Weeratna RD, McCluskie MJ, Davis HL. Immune-mediated destruction of transfected myocytes following DNA vaccination occurs via multiple mechanisms. Gene Ther. 2001; 8: 1395–1400.
- You Z, Huang X, Hester J, Toh HC, Chen SY. Targeting dendritic cells to enhance DNA vaccine potency. Cancer Res. 2001; 61: 3704–3711.
- Klinman DM, Klaschik S, Tross D, Shirota H, Steinhagen F. FDA guidance on prophylactic DNA vaccines: analysis and recommendations. Vaccine. 2010; 28: 2801–2805.
- Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr Gene Ther. 2002; 2: 183–194.
- Bolhassani A, Safaiyan S, Rafati S. Improvement of different vaccine delivery systems for cancer therapy. Mol Cancer. 2011; 10: 3.
- Pichon C, Billiet L, Midoux P. Chemical vectors for gene delivery: uptake and intracellular trafficking. Curr Opin Biotechnol. 2010; 21: 640–645.
- Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005 Apr; 11: S45–53.
- Schoen C, Stritzker J, Goebel W, Pilgrim S. Bacteria as DNA vaccine carriers for genetic immunization. Int J Med Microbiol IJMM. 2004; 294: 319–335.
- Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004; 5: 971–974.
- Seow Y, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Mol Ther J Am Soc Gene Ther. 2009; 17: 767–777.
- Srivastava IK, Liu MA. Gene vaccines. Ann Intern Med. 2003; 138: 550–559.
- Mitragotri S. Immunization without needles. Nat Rev Immunol. 2005; 5: 905–16.
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001; 2: 361–367.
- Grillot-Courvalin C, Goussard S, Courvalin P. Bacteria as gene delivery vectors for mammalian cells. Curr Opin Biotechnol. 1999; 10: 477–481.
- Saha R, Killian S, Donofrio RS. DNA vaccines: a mini review. Recent Pat DNA Gene Seq. 2011; 5: 92–96.
- Barbosa T, Rescigno M. Host-bacteria interactions in the intestine: homeostasis to chronic inflammation. Wiley Interdiscip Rev Syst Biol Med. 2010; 2: 80–97.
- Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine. 2011; 29: 3341–3355.
- Gurunathan S, Wu CY, Freidag BL, Seder RA. DNA vaccines: a key for inducing long-term cellular immunity. Curr Opin Immunol. 2000; 12: 442–447.
- Sheikh NA, Morrow WJW. Guns, genes, and spleen: a coming of age for rational vaccine design. Methods San Diego Calif. 2003; 31: 183–192.
- Brandtzaeg P, Kiyono H, Pabst R, Russell MW. Terminology: nomenclature of mucosa-associated lymphoid tissue. Mucosal Immunol. 2008; 1: 31–37.
- Rescigno M. Gut commensal flora: tolerance and homeostasis. F1000 Biol Rep [Internet]. 2009; 1.
- Jung C, Hugot J-P, Barreau F. Peyer’s Patches: The Immune Sensors of the Intestine. Int J Inflamm [Internet]. 2010; 2010.
- Kosiewicz MM, Zirnheld AL, Alard P. Gut Microbiota, Immunity, and Disease: A Complex Relationship. Front Microbiol [Internet]. 2011; 2.
- Yu X, Jia R, Huang J, Shu B, Zhu D, Liu Q, et al. Attenuated Salmonella typhimurium delivering DNA vaccine encoding duck enteritis virus UL24 induced systemic and mucosal immune responses and conferred good protection against challenge. Vet Res. 2012; 43: 56.
- Wen J, Yang Y, Zhao G, Tong S, Yu H, Jin X, et al. Salmonella typhi Ty21a bacterial ghost vector augments HIV-1 gp140 DNA vaccine-induced peripheral and mucosal antibody responses via TLR4 pathway. Vaccine. 2012; 30: 5733–5739.
- Al-Mariri A, Tibor A, Lestrate P, Mertens P, De Bolle X, Letesson J-J. Yersinia enterocolitica as a Vehicle for a Naked DNA Vaccine Encoding Brucella abortus Bacterioferritin or P39 Antigen. Infect Immun. 2002; 70: 1915–1923.
- Tangney M, Gahan CGM. Listeria monocytogenes as a vector for anti-cancer therapies. Curr Gene Ther. 2010; 10: 46–55.
- Shahabi V, Reyes-Reyes M, Wallecha A, Rivera S, Paterson Y, Maciag P. Development of a Listeria monocytogenes based vaccine against prostate cancer. Cancer Immunol Immunother CII. 2008; 57: 1301–1313.
- Minor PD, Dunn G. The effect of sequences in the 5’ non-coding region on the replication of polioviruses in the human gut. J Gen Virol. 1988; 69: 1091–1096.
- Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol. 2008; 6: 349–362.
- Steidler L, Rottiers P. Therapeutic drug delivery by genetically modified Lactococcus lactis. Ann N Y Acad Sci. 2006; 1072: 176–186.
- Carr FJ, Chill D, Maida N. The Lactic Acid Bacteria: A Literature Survey. Crit Rev Microbiol. 2002; 28: 281–370.
- Price CE, Zeyniyev A, Kuipers OP, Kok J. From meadows to milk to mucosa - adaptation of Streptococcus and Lactococcus species to their nutritional environments. FEMS Microbiol Rev. 2012; 36: 949–971.
- Prakash S, Tomaro-Duchesneau C, Saha S, Cantor A. The Gut Microbiota and Human Health with an Emphasis on the Use of Microencapsulated Bacterial Cells. BioMed Res Int. 2011; 2011: e981214.
- Van de Guchte M, Ehrlich SD, Maguin E. Production of growth-inhibiting factors by Lactobacillus delbrueckii. J Appl Microbiol. 2001; 91: 147–153.
- Food and Agriculture Organization of the United Nations, World Health Organization, editors. Probiotics in food: health and nutritional properties and guidelines for evaluation. Rome: Food and Agriculture Organization of the United Nations : World Health Organization. 2006; 50.
- Masood MI, Qadir MI, Shirazi JH, Khan IU. Beneficial effects of lactic acid bacteria on human beings. Crit Rev Microbiol. 2011; 37: 91–98.
- Fuller R, Gibson GR. Modification of the intestinal microflora using probiotics and prebiotics. Scand J Gastroenterol Suppl. 1997; 222: 28–31.
- Bolotin A, Mauger S, Malarme K, Ehrlich SD, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Van Leeuwenhoek. 1999; 76: 27–76.
- De Vos WM. Gene expression systems for lactic acid bacteria. Curr Opin Microbiol. 1999; 2: 289–295.
- Vos WMD, Simons GFM. Gene cloning and expression systems in Lactococci. In: Gasson MJ, Vos WMD, editors. Genetics and Biotechnology of Lactic Acid Bacteria [Internet]. Springer Netherlands; 1994. p. 52–105.
- Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001; 11: 731–753.
- Van Asseldonk M, Rutten G, Oteman M, Siezen RJ, de Vos WM, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990; 95: 155–160.
- Loir YL, Azevedo V, Oliveira SC, Freitas DA, Miyoshi A, Bermúdez-Humarán LG, et al. Protein secretion in Lactococcus lactis : an efficient way to increase the overall heterologous protein production. Microb Cell Factories. 2005; 4: 2.
- Wells JM, Wilson PW, Norton PM, Gasson MJ, Le Page RW. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993 Jun;8: 1155–1162.
- Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem FEBS. 1993; 216: 281–291.
- Bredmose L, Madsen SM, Vrang A, Ravn P, Johnsen MG, Glenting J, et al. Development of a Heterologous Gene Expression System for Use in Lactococcus Lactis. In: Merten O-W, Mattanovich D, Lang C, Larsson G, Neubauer P, Porro D, et al., editors. Recombinant Protein Production with Prokaryotic and Eukaryotic Cells A Comparative View on Host Physiology [Internet]. Springer Netherlands; 2001. p. 269–275.
- Madsen SM, Arnau J, Vrang A, Givskov M, Israelsen H. Molecular characterization of the pH-inducible and growth phase-dependent promoter P170 of Lactococcus lactis. Mol Microbiol. 1999; 32: 75–87.
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983; 22: 181–189.
- Miyoshi A, Jamet E, Commissaire J, Renault P, Langella P, Azevedo V. A xylose-inducible expression system for Lactococcus lactis. FEMS Microbiol Lett. 2004; 239: 205–212.
- Llull D, Poquet I. New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl Environ Microbiol. 2004; 70: 5398–5406.
- Pontes DS, de Azevedo MSP, Chatel J-M, Langella P, Azevedo V, Miyoshi A. Lactococcus lactis as a live vector: heterologous protein production and DNA delivery systems. Protein Expr Purif. 2011; 79: 165–175.
- Bermúdez-Humarán LG, Innocentin S, Lefèvre F, Chatel J-M, Langella P. Development of Mucosal Vaccines Based on Lactic Acid Bacteria. In: Charalampopoulos D, Rastall RA, editors. Prebiotics and Probiotics Science and Technology [Internet]. Springer New York; 2009. p. 1099–1122.
- Kleerebezem M, Beerthuyzen MM, Vaughan EE, de Vos WM, Kuipers OP. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997; 63: 4581–4584.
- Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005; 68: 705–717.
- De Ruyter PG, Kuipers OP, de Vos WM. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996; 62: 3662–3667.
- Almeida JF, Mariat D, Azevedo V, Miyoshi A, de Moreno de LeBlanc A, Del Carmen S, et al. Correlation between fibronectin binding protein A expression level at the surface of recombinant lactococcus lactis and plasmid transfer in vitro and in vivo. BMC Microbiol. 2014; 14: 248.
- Eichenbaum Z, Federle MJ, Marra D, de Vos WM, Kuipers OP, Kleerebezem M, et al. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol. 1998; 64: 2763–2769.
- Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, et al. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl Environ Microbiol. 2000; 66: 4427–4432.
- Lokman BC, Leer RJ, van Sorge R, Pouwels PH. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol Gen Genet MGG. 1994; 245: 117–125.
- Jamet E, Renault P. Etude de l’expression et de la régulation des gènes impliqués dans le métabolisme carboné chez Lactococcus lactis = Expression and regulation study of genes involved in carbon metabolism in Lactococcus lactis [Internet]. 2001.
- Azevedo M de, Karczewski J, Lefévre F, Azevedo V, Miyoshi A, Wells JM, et al. In vitro and in vivo characterization of DNA delivery using recombinant Lactococcus lactis expressing a mutated form of L. monocytogenes Internalin A. BMC Microbiol. 2012; 12: 299.
- Hollmann A, Saviello M, Delfederico L, Saraiva TDL, Barh D, Jain N, et al. Tight controlled expression and secretion of Lactobacillus brevis SlpA in Lactococcus lactis. Biotechnol Lett. 2012; 34: 1275–1281.
- Le Loir Y, Nouaille S, Commissaire J, Brétigny L, Gruss A, Langella P. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microbiol. 2001; 67: 4119–4127.
- Chatel J-M, Pothelune L, Ah-Leung S, Corthier G, Wal J-M, Langella P. In vivo transfer of plasmid from food-grade transiting lactococci to murine epithelial cells. Gene Ther. 2008; 15: 1184–1190.
- Guimarães VD, Gabriel JE, Lefèvre F, Cabanes D, Gruss A, Cossart P, et al. Internalin-expressing Lactococcus lactis is able to invade small intestine of guinea pigs and deliver DNA into mammalian epithelial cells. Microbes Infect Inst Pasteur. 2005; 7: 836–844.
- Innocentin S, Guimarães V, Miyoshi A, Azevedo V, Langella P, Chatel J-M, et al. Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl Environ Microbiol. 2009; 75: 4870–4878.
- Robinson K, Chamberlain LM, Schofield KM, Wells JM, Le Page RW. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat Biotechnol. 1997; 15: 653–657.
- Chang TL-Y, Chang C-H, Simpson DA, Xu Q, Martin PK, Lagenaur LA, et al. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc Natl Acad Sci U S A. 2003; 100: 11672–11677.
- Guimarães VD, Innocentin S, Lefèvre F, Azevedo V, Wal J-M, Langella P, et al. Use of native lactococci as vehicles for delivery of DNA into mammalian epithelial cells. Appl Environ Microbiol. 2006; 72: 7091–7097.
- Salminen S, von Wright A, Morelli L, Marteau P, Brassart D, de Vos WM, et al. Demonstration of safety of probiotics -- a review. Int J Food Microbiol. 1998; 44: 93–106.
- Neutra MR, Kozlowski PA. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol. 2006; 6: 148–158.
- Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast Chichester Engl. 2005; 22: 249–270.
- Wollert T, Pasche B, Rochon M, Deppenmeier S, van den Heuvel J, Gruber AD, et al. Extending the host range of Listeria monocytogenes by rational protein design. Cell. 2007; 129: 891–902.
- Que YA, François P, Haefliger JA, Entenza JM, Vaudaux P, Moreillon P. Reassessing the role of Staphylococcus aureus clumping factor and fibronectin-binding protein by expression in Lactococcus lactis. Infect Immun. 2001; 69: 6296–6302.
- Guimarães V, Innocentin S, Chatel J-M, Lefèvre F, Langella P, Azevedo V, et al. A new plasmid vector for DNA delivery using lactococci. Genet Vaccines Ther. 2009; 7: 4.
- Pontes D, Azevedo M, Innocentin S, Blugeon S, Lefévre F, Azevedo V, et al. Immune response elicited by DNA vaccination using Lactococcus lactis is modified by the production of surface exposed pathogenic protein. PloS One. 2014; 9: e84509.
- Del Carmen S, Martín Rosique R, Saraiva T, Zurita-Turk M, Miyoshi A, Azevedo V, et al. Protective effects of lactococci strains delivering either IL-10 protein or cDNA in a TNBS-induced chronic colitis model. J Clin Gastroenterol. 2014; 48: S12–17.
- Sim ACN, Lin W, Tan GKX, Sim MST, Chow VTK, Alonso S. Induction of neutralizing antibodies against dengue virus type 2 upon mucosal administration of a recombinant Lactococcus lactis strain expressing envelope domain III antigen. Vaccine. 2008; 26: 1145–1154.
- Xin K-Q, Hoshino Y, Toda Y, Igimi S, Kojima Y, Jounai N, et al. Immunogenicity and protective efficacy of orally administered recombinant Lactococcus lactis expressing surface-bound HIV Env. Blood. 2003; 102: 223–228.
- Scavone P, Miyoshi A, Rial A, Chabalgoity A, Langella P, Azevedo V, et al. Intranasal immunisation with recombinant Lactococcus lactis displaying either anchored or secreted forms of Proteus mirabilis MrpA fimbrial protein confers specific immune response and induces a significant reduction of kidney bacterial colonisation in mice. Microbes Infect Inst Pasteur. 2007; 9: 821–828.
- Hanniffy SB, Carter AT, Hitchin E, Wells JM. Mucosal delivery of a pneumococcal vaccine using Lactococcus lactis affords protection against respiratory infection. J Infect Dis. 2007; 195: 185–193.
- Daniel C, Sebbane F, Poiret S, Goudercourt D, Dewulf J, Mullet C, et al. Protection against Yersinia pseudotuberculosis infection conferred by a Lactococcus lactis mucosal delivery vector secreting LcrV. Vaccine. 2009; 27: 1141–1144.
- Ahmed B, Loos M, Vanrompay D, Cox E. Mucosal priming of the murine immune system against enterohemorrhagic Escherichia coli O157:H7 using Lactococcus lactis expressing the type III secretion system protein EspB. Vet Immunol Immunopathol. 2013; 152: 141–145.
- Hugentobler F, Di Roberto RB, Gillard J, Cousineau B. Oral immunization using live Lactococcus lactis co-expressing LACK and IL-12 protects BALB/c mice against Leishmania major infection. Vaccine. 2012; 30: 5726–5732.
- Bermúdez-Humarán LG, Cortes-Perez NG, L’Haridon R, Langella P. Production of biological active murine IFN-gamma by recombinant Lactococcus lactis. FEMS Microbiol Lett. 2008; 280: 144–149.
- Zamri HF, Shamsudin MN, Rahim RA, Neela V. Oral vaccination with Lactococcus lactis expressing the Vibrio cholerae Wzm protein to enhance mucosal and systemic immunity. Vaccine. 2012; 30: 3231–3238.
- De Moreno de LeBlanc A, LeBlanc JG, Perdigón G, Miyoshi A, Langella P, Azevedo V, et al. Oral administration of a catalase-producing Lactococcus lactis can prevent a chemically induced colon cancer in mice. J Med Microbiol. 2008; 57: 100–105.
- Cortes-Perez NG, Lefèvre F, Corthier G, Adel-Patient K, Langella P, Bermúdez-Humarán LG. Influence of the route of immunization and the nature of the bacterial vector on immunogenicity of mucosal vaccines based on lactic acid bacteria. Vaccine. 2007; 25: 6581–6588.
- Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon J-P, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2006; 4: 754–759.
- Marinho F a. V, Pacífico LGG, Miyoshi A, Azevedo V, Le Loir Y, Guimarães VD, et al. An intranasal administration of Lactococcus lactis strains expressing recombinant interleukin-10 modulates acute allergic airway inflammation in a murine model. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2010; 40: 1541–1551.