
Research Article
Austin J Vet Sci & Anim Husb. 2019; 6(3): 1063.
Prevalence of Escherichia coli Plasmid mcr-1 in Rabbits in Shandong, China
Wang X1,2, Hu D1, Zhai Z3, Zhao X4, Zhang H5, Jiang H6, Yi S6, Peng J1, Zhai J6* and Chang W1*
1College of Animal Science and Technology, Shandong Agricultural University, Taian, China
2School of Public Health, Shandong First Medical University & Shandong Academy of Medical Sciences, Taian, China
3Postdoctoral Scientific Research Station, Taian City Central Hospital, Taian, China
4Institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences, Ji’nan, China
5Department of Teaching Affairs, Hebei University of Economics and Business, Shijiazhuang, China
6College of Basic Medicine, Shandong First Medical University & Shandong Academy of Medical Sciences, Taian, China
*Corresponding author: Zhai J, College of Animal Science and Technology, Shandong Agricultural University, Taian, China
Chang W, College of Basic Medicine, Shandong First Medical University & Shandong Academy of Medical Sciences, Taian, China
Received: September 30, 2019; Accepted: November 01, 2019; Published: November 08, 2019
Abstract
The antibiotic resistance gene mcr-1 is widespread in domestic and wild animals. Therefore, continuous monitoring of its prevalence and characteristics is required. In this study, we applied a PCR-based method to detect mcr- 1 of Escherichia coli in rabbits of Tai’an, China. A total of 55 non-duplicated E. coli samples were recovered from the swabs of rabbit feces. Plasmid and chromosome PCR, a conjugation experiment, lactose fermentation experiment, multilocus sequence typing, and antibiotic resistance tests were performed to determine the characteristics of mcr-1-bearing plasmids. Bacterial plasmids and chromosome DNA were separately extracted and amplified by PCR with mcr-1-specific primers. Eight of the 55 specimens were mcr-1-positive, for a positive rate of 14.6%. The mcr-1-positive E. coli harbored more drug-resistant genes compared with the mcr-1-negative specimens and results showed diverse sequence types. All the E. coli isolates were sensitive to ceftazidime, ceftriaxone, imipenem and amikacin while 14.6% of the isolates showed resistance against polymycin and 65.5% were resistant against ampicillin. Although mcr-1 was successfully amplified with PCR from bacterial plasmids, it could not be amplified from bacterial chromosome DNA. Overall, mcr-1 has been first isolated from rabbits, and these findings suggest the possible threat of the transmission of mcr-1 from rabbits to humans, primarily since the gene is located on transferable plasmids making horizontal transfer relatively easy. Since food-producing animals are necessary for our daily diet, global cooperation is needed in fighting the spread of this drug resistance gene to avoid human infections with multidrug-resistant pathogenic bacteria.
Keywords: Rabbits; mcr-1; Plasmids; Escherichia coli; Shandong Province
Novelty Statement: mCR-1 gene is a polymycin-resistant gene discovered in recent years, which is an important discovery in the world. After that, mcr-1 were found in Escherichia coli and Klebsiella pneumoniae in chickens, ducks, geese, pigs, and other omnivorous animals. Rabbits belong to herbivores, and this is the first time that mcr-1 has been found in rabbits in China.
Introduction
With the widespread use of antibiotics in farming, drug-resistant genes are now widely distributed in the intestines of farm animals, which are continuously being identified [1-3]. Following this pattern, it is likely that drug-resistant bacteria are present in rabbit feces [4,5]. Furthermore, antimicrobial resistant bacteria can be transferred to the humans through the food chain, thus affecting human health. Therefore, in the present study, we employed a simpler and more economical method to determine the location and characteristics of E. coli mcr-1 among rabbits in China. We also developed a method of combination of PCR and lactose fermination test to prove further that the plasmid is harboring mcr-1. For the final determination, we applied plasmid whole genome sequencing to the mcr-1 positive strains.
Polymyxin is a promising antimicrobial peptide, and very few bacteria show polymyxin resistance at present. However, Chinese researchers recently identified mcr-1 as a gene conferring resistance to colistin and polymyxin [6,7]. Although mcr-1 has been reported and detected worldwide, its global prevalence remains mostly unknown. Liu et al. screened for mcr-1 in Escherichia coli in raw pork and found that the gene was located on a plasmid. The prevalence of Escherichia coli mcr-1 in rabbits in China has not been reported. In these studies, the key methods to detect the location of genes were based on Southern blotting or whole genome sequencing. However, their detection methods were not based on Polymerase Chain Reaction (PCR) amplification, which can help in estimating the prevalence of mcr-1.
Materials and Methods
Sample collection and identification of E. coli
The rabbits had been raised on large rabbit farms free from thirst or starvation. The formula for rabbit feed is 17% corn, 24% bran, 21% soybean meal, 5% imported fish meal, 3% active yeast and 30% grass powder. Fecal samples were randomly collected from the diarrhea of rabbits on three farms. Because the sampling process did not harm the rabbits, ethical approval was not required for the study.
Rabbit feces were collected in aseptic tubes [8] and plated on MacConkey agar as well as placed in micro chemical tubes to select and identify E. coli. The suspicious colonies were identified by bacterial biochemical tests (Table 1).
Test item
Test result
Test item
Test result
Sucrose
Positive
M-R Test
Positive
Lactose
Positive
V-P Test
Negative
Glucose
Positive
H2S Test
Negative
Maltose
Positive
Indole Test
Positive
Mannitol
Positive
Table 1: Biochemical test results of E. coli strains.
PCR detection of mcr-1
The DNA from 55 E. coli strains was amplified by PCR with mcr- 1 whole sequence specific primers. The PCR products of mcr-1 were then subjected to electrophoresis. The positive specimens were sent to Sangon for direct sequencing for confirmation [9], and the sequences of mcr-1-positive strains were compared by the Blastn tool of the National Center for Biotechnology Information website.
We further attempted to amplify the mcr-1 gene from extracted plasmids and bacterial chromosomes, respectively. PCR was then carried out with the extracted plasmid as a template using primers specific to mcr-1 and other resistance genes under the reference PCR conditions described above (Supplementary Table S1). Similarly, bacterial genomic chromosomes were extracted and purified from the samples of mcr-1-positive strains, and PCR analysis was performed with mcr-1-specific primers as described above.
Plasmid characterization and sequencing
Those plasmids meeting the requirements of sequencing were sent to Shanghai Pinoson Biological Co., Ltd. for whole-genome sequencing, and the coding genes and structure were analyzed by bioinformatics. A whole-genome shotgun strategy was used to construct libraries of different inserted fragments. Paired-end sequencing was performed on the Illumina MiSeq platform. Finally, a complete plasmid sequence was obtained by assembling overlapping groups and filling vacancy sequences by a combinatorial PCR or stepby- step method.
Conjugation experiments
The transferability of mcr-1-bearing plasmids from isolates was determined using filter mating with E. coli J53 as the recipient strain, mixing at a ratio of 1:1 in broth culture, as previously described [10]. The resulting transconjugants were selected on brain heart infusion agar plates supplemented with polymyxin B (2mg/L) [11]. Subsequently, the positive bacteria were cultivated together with the mcr-1-negative receptor J53, which contained no plasmid. The conjugated bacteria were observed using plasmid extraction and electrophoresis analysis.
Conjugated strains of E. coli were also subjected to a lactose fermentation experiment, with the mcr-1-positive strain J53 as the negative control.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of the E. coli isolates was tested according to the determination of the Minimal Inhibitory Concentration (MIC) of several antibiotics. The susceptibility of the isolates was then tested with 15 kinds of commonly used antimicrobial agents. E. coli isolates resistant to more than three classes of antimicrobials were defined as Multidrug-Resistant (MDR) isolates [12,13].
Multilocus sequence typing (MLST)
According to Zhao et al. eight pairs of primers for housekeeping genes were designed and used for PCR. The products of PCR amplification were then sequenced by Shanghai Sangon Biotech Co., Ltd. The results were amended using Chromas and DNA Star software and then submitted to the Pasteur online database for processing Zhao et al. The allele number of each housekeeping gene was obtained and the Sequence Type (ST) of each strain was acquired [14].
Results
Prevalence of mcr-1
Eight of the 55 specimens were found to be mcr-1-positive, representing a positivity rate of 14.6% (Figure 1). Although mcr-1 was successfully PCR-amplified from bacterial plasmids, it could not be amplified from bacterial chromosome DNA, suggesting that the mcr-1 resistant gene may locate on the plasmid and not on genomic chromosomes.

Figure 1: mcr-1-positive E. coli.
The mcr-1-positive strains harbored significantly more drugresistant genes other than mcr-1 compared to the mcr-1-negative strains (chi square test, P‹0.05; Table 2). Accordingly, the mcr-1- positive E. coli had a greater probability of being MDR than mcr-1 negative E. coli (P‹0.05).
Yes
No
MDR
No.
Rate
No.
Rate
mcr-1 Positive
7
87.50%
1
12.50%
mcr-1 Negative
23
48.94%
24
51.06%
Table 2: Comparison of multiple drug-resistant isolates detected in mcr-1- positive and -negative strains.
Plasmid sequencing results
Plasmid whole-genome sequencing was conducted on the mcr-1 positive strains. Blastn showed that mcr-1 was located on the plasmid. The extracted plasmid, designated pR45, was found to be a closed-loop DNA molecule with 83,157 bp and a 52.74% GC content, encoding 45 predicted genes, including four known resistance genes: mcr-1, blaCTX-M, blaTEM-1, and qnrS1. To prove the transferability of mobile plasmids in vitro, E. coli strain R45, carrying the mcr-1, blaCTX-M, blaTEM-1, and qnrS1 genes, was selected for comparing and analyzing the extracted plasmids. The results of drug resistance phenotyping and resistance gene detection of conjugated bacteria in vitro were consistent with the results of plasmid sequencing, demonstrating that the E. coli resistance gene has transferability in vitro, and that the mobile plasmid plays an essential role in the process of drug resistance transmission in E. coli.
Conjugation tests
The conjugation tests confirmed the horizontal transfer of mcr-1 in E. coli strains obtained from rabbit feces, therefore proving that mcr-1 was located on plasmids. The mcr-1-positive bacteria were then cultivated together with the mcr-1-negative strain J53, which contained no plasmid. The transfer of the resistance gene was found to take place when the workable plasmid was transferred from the wild type mcr-1 positive bacterium to the recipient. Moreover, the conjugated bacteria acquired lactose fermentation ability and showed an increase in polymyxin resistance ability (Figure 2, Figure 3, Table 4).
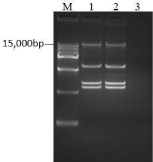
Figure 2: Identical plasmid profile of the donor and conjugant.

Figure 3: Drug sensitivity tests.
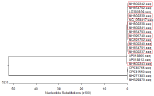
Figure 4: Phylogenetic tree of mcr-1 detected in E. coli isolated from rabbits.
In addition, the plasmid DNA of mcr-1-positive E. coli strongly amplified mcr-1. The target band was purified and subjected to PCR detection using primers for both mcr-1 and blaTEM, which showed positive results indicating the two resistant genes coexist on the same plasmid. BlaTEM was included in this analysis as it is the most common AMR genes in the samples, with a positive rate of 98.2%.
Characteristics of mcr-1
Thirteen different STs were identified among the 55 strains, with the most prevalent being ST302 (22/55, 40.0%), ST370 (12/55, 21.8%), and ST468 (Table 3). Of note, the mcr-1-positive E. coli strains also showed a wide diversity of STs, although the dominant type was ST88 (62.5%).
Strain
Genbank
ST
Resistance phenotype
Resistance
R45
MH602237
ST88
AML-AMP-C-CIP-GEN- NA-SXT-TET-PB
blaCTX-M, blaTEM, cmlA, flor, sul2, sul3, tetB, mcr-1
R48
MH602238
ST88
AMP-C-CIP-GEN-NA-SXT-TET-PB
blaCTX-M, blaTEM,cmlA, flor, sul2, sul3, tetB, mcr-1
R49
MH602239
ST2
AMP-C-CIP-NA-SXT-TET-PB
blaCTX-M, blaTEM, cmlA, flor, sul3, mcr-1
R50
MH602240
ST88
AMP-C-CIP-GEN-NA-SXT-TET-PB
blaCTX-M, blaTEM, cmlA, flor, sul2, sul3, tetB, mcr-1
R51
MH602241
ST353
C-TET-PB
blaTEM, flor, qnrS, sul2, mcr-1
R54
MH602242
ST88
C-CIP-NA-TET-PB
blaCTX-M, blaTEM, flor, sul2, sul3, tetB, mcr-1
R54
MH602243
ST24
AML-AMP-TET-PB
blaCTX-M, blaTEM, flor, sul1, mcr-1
R55
MH395740
ST88
AMP-C-CIP-GEN-NA-SXT-TB-TET-PB
blaCTX-M, blaTEM, cmlA, flor, sul2, sul3, tetB, mcr-1
Table 3: Characteristics of mcr-1 of E. coli in rabbits.
Lactose fermentation
Plasmid
mcr-1 (Plasmid)
donor
+ (yellow)
+
+
recipient
- (purple)
-
-
zygote
+ (yellow)
+
+
Table 4: Lactose fermentation results.
Figure 5 shows the phylogenetic tree to display the evolutionary relationships among the eight mcr-1 sequences, demonstrating that although the eight positive strains were non-duplicated E. coli, their mcr-1 sequences were very similar.
Discussion
Prevalence of mcr-1 in E. coli
The prevalence of mcr-1 (8/55, 14.6%) detected in E. coli strains obtained from rabbits in Tai’an, China is similar to that reported in a study conducted in Italy (50/320, 15.6%) Fabrizio et al. and is markedly higher than that reported for humans (1~2%) [15]. This high rate may be due to the greater use of polymyxin in farms than in clinical practice. More importantly, all of the mcr-1-positive strains obtained in the present study were isolated from a single farm among the three sampled farms. This may be related to several factors. First, the sample size might not have been large enough to reflect the actual situation at all farms. Second, the horizontal transfer of mcr-1 was confined within each relatively closed farm, thereby preventing gene transfer among farms, especially farms from different regions. Finally, but potentially most important, the amount of polymyxin use varied across the different farms, which would impose different selection pressures on mcr-1. Of note, mcr-1 has been found in lots of animals such as livestock and poultry, birds, and wild animals. As far as we know, this is the first time that mcr-1 was isolated from rabbits.
This study showed that E. coli isolated from diarrheic farmed rabbits in the Tai’an area exhibit sometimes very frequent resistance to antimicrobials important to human medicine, which further highlights the need for reasonable use of antibiotics. In this study, the highest isolation rate of 40% (22/55) was found for ST302 which had not yet been reported relating to infections, while the highest isolation rate of 37.5% (3/8) was found for ST88 among mcr-1 positive strains.
Dissemination characteristics of mcr-1
Because of the limitation of the total number of specimens, it is difficult to generalize the results overall. Nevertheless, the antibiotic resistance tests demonstrated that the mcr-1-positive plasmids were more likely to harbor other resistant genes than the mcr-1-negative plasmid (Table 2). Bacteria without plasmids readily gained donor bacterium plasmids and the mcr-1 gene along with the ability for lactose fermentation and polymyxin resistance at the same time. Therefore, these results strongly suggest the high horizontal dissemination potential of mcr-1.
Moreover, the low diversity of mcr-1 sequences among the E. coli strains from different sources indicated that the mcr-1 gene was most likely derived from one ancestor, further suggesting clonal transmission of E. coli and horizontal transmission of mcr-1-bearing plasmids in this area. This may be related to the fact that this region is relatively isolated, far from the city, with a minimal flow of people. Additionally, the rabbit feed contains the same fish meal, which may contain mcr-1 positive bacteria and thus infect the rabbit when eaten.
The resistance gene mcr-1 was found in eight strains of bacteria, which shows that the presence of plasmids for bacteria makes it possible to produce drug resistance and survive in adversity [16,17]. Resistance genes not only transfer from one bacterium to another or from one bacterium specy to other species but also move geographically consequencely. Therefore, the threat of drug resistance is not localized to a given animal farm or region but represents a worldwide concern requiring global cooperation. Indeed, the fact that the bacterial resistant gene is located on the plasmid makes it potentially more difficult to control than a chromosomal gene. Plasmid transmission makes the spread of drug resistance genes easier and faster, and since the same plasmid can carry a variety of resistance genes, the recipient can immediately become resistant to multiple drugs. This finding suggests that it would be very challenging to cure humans infected with multiple drug-resistant pathogenic bacteria.
Conclusion
The conjugation test and whole-genome sequence analysis of the ligated plasmid demonstrated that the E. coli resistance gene mcr- 1 is circulating in rabbits of China, with the ability for horizontal transfer in vitro, indicating that the mobile plasmid plays a vital role in the process of antibiotic resistance of E. coli. As the AMR positive bacterial strains can survive in the presence of antibiotics, they may acquire additional drug resistance genes, resulting in a new MDR phenotype for the donor bacteria. Therefore, the continuous selective pressure of antibiotics in farms will result in the production of new drug resistance genes that can readily circulate among domestic and wild animals, and even humans.
Funding
This work was supported by the Key R & D projects in Shandong (2015GSF119023), the National Natural Science Fund (NSFC81401328, and Shandong Provincial Natural Science Foundation, China (ZR2013HL064).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Briñas L, Zarazaga M, Saenz Y, Ruiz-Larrea F, Torres C. Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 2002; 46: 3156-3163.
- Naseer U, Sundsfjord A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 2011; 17: 83-97.
- Bryan A, Shapir N, Sadowsky MJ. Frequency and distribution of tetracycline resistance genes in genetically diverse, non-selected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 2004; 70: 2503-2507.
- Zhao X, Ye C, Chang W, Sun S. Serotype distribution, antimicrobial resistance, and class I integrons profiles of Salmonella from animals in slaughterhouses in Shandong Province, China. Front. Microbiol. 2017; 8: 1-9.
- Gao LL, Hu, JQ, Zhang, XD, Wei L, Li S, Miao Z, et al. Application of swine manure on agricultural fields contributes to extended-spectrum β-lactamaseproducing Escherichia coli spread in Tai’an, China. Front. Microbiol. 2015; 6: 313.
- Agnoletti F, Brunetta R, Bano L, Drigo I, Mazzolini E. Longitudinal study on antimicrobial consumption and resistance in rabbit farming. Int. J. Antimicrob. Agents. 2018; 51: 197-205.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016; 16: 161-168.
- Agnoletti F. Update on rabbit enteric diseases: despite improved diagnostic capacity, where does disease control and prevention stand? Proc. “10th World Rabbit Congr”, Sharm El-Sheikh, Egypt. 2012; 113-1127.
- White P, Mclver CJ, Deng YM, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 2006; 182: 265- 269.
- Zhang H, Zhou Y, Guo S, Chang W. Multidrug resistance found in extendedspectrum beta-lactamase-producing Enterobacteriaceae from rural water reservoirs in Guantao, China. Front. Microbiol. 2015; 6: 1-4.
- Guardabassi L, Schwarz SD, Lloyd H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004; 54: 321- 332.
- Valat C, Forest K, Billet M, Polizzi C, Saras E, Madec JY, et al. Absence of co-localization between pathovar-associated virulence factors and extendedspectrum β-lactamase (blaCTX-M) genes on a single plasmid. Vet. Microbiol. 2016; 192: 163-166.
- Moawad AA, Hotzel H, Awad O, Tomaso H, Neubauer H, Hafez HM, et al. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017; 9: 57.
- Dotto G, Giacomell M, Grilli G, Ferrazzi V, Carattoli A, Fortini D, et al. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb. Drug Resist. 2014; 20: 118-123.
- Yi LX, Liu YY, Wu RJ, Liang ZS, Liu JH. Research progress on the plasmidmediated colistin resistance gene mcr-1. Yi Chuan. 2017; 39: 110-126.
- Kun QF, Geng Y, Yu ZH, et al. Detection of quinolone resistance and plasmid mediated quinolone gene in Escherichia coli isolated from rabbit. Chinese J. Prevent. Vet. Med. 2006; 38: 944-948.
- Silva N, Igrejas G, Figueredo N, Gonçalves A, Radhouani H, Rodrigues J, et al. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010; 408: 4871-4876.