
Research Article
Austin J Vet Sci & Anim Husb. 2022; 9(1): 1089.
Isolation and Molecular Detection of Lumpy Skin Disease Virus from Outbreak Cases in Illubabor Zone, Oromia Regional State, Ethiopia
Guyassa C¹*, Dilbato T², Aliy A¹, Shegu D¹ and Zewude D¹
¹National Animal Health Diagnostic and Investigation Center, Sebeta, Ethiopia
²Ambo University, College Of Agriculture and Veterinary Science, Ambo, Ethiopia
*Corresponding author: Chala Guyassa, National Animal Health Diagnostic and Investigation Center, Sebeta, Ethiopia
Received: February 17, 2022; Accepted: March 11, 2022; Published: March 18, 2022
Abstract
Background: Lumpy skin disease is one of the most economically important viral diseases of cattle and Asian water buffalo caused by lumpy skin disease virus, which occurs in most African countries including Ethiopia. In Ethiopia, it has been detected in exotic and local breed cattle. To this end, there is scarcity of information on its epidemic status, among cattle in Illubabor, Ethiopia. Therefore, outbreak investigation was conducted to estimate epidemic status of lumpy skin disease among cattle in illubabor zone, Yayo district.
Methodology: Outbreak investigation was done from August to December 2020 in the Ilubabor zone, Yayo district, with the goal of isolating and detecting the virus using molecular methods. All ages and both sexes of local breed cattle from reported disease outbreaks of the study area were subjected to the study. Skin biopsies (n=44) were collected from non-vaccinated lumpy skin disease affected cattle after examining the presence of skin lesions and transported to NAHDIC for laboratory test. Virus was isolated by growing on Vero cells and molecular detection was performed by conventional and real-time polymerase chain reaction.
Results: The characteristic capripoxvirus cytopathic effect was observed on 17 out of 44 Vero cells inoculated. Out of the total 44 samples, 88.63% (39/44) and (95.45%) 42/44 were found positive for LSDV by conventional and real time polymerase chain reaction respectively. Chi-square (x²) test was used to assess the association of sex and ages with affected group. Morbidity, mortality and case fatality rates were 15.49%, 1.4% and 9.09% respectively. Adult cattle showed higher morbidity (17.18%) than young ones (11.1%). Also not statistically significant, higher morbidity was observed in female (16.66%) than male (14.11%) cattle.
Conclusion: The study showed that lumpy skin disease was circulating in cattle in the area and causing great loss to the farmers with high morbidity rates. To reduce economic losses caused by the disease, it was suggested that strategic programs for effective control and prevention be established.
Keywords: Illubabor Zone; Lumpy skin disease virus; Molecular detection; Polymerase chain reaction; Virus isolation
Introduction
Ethiopia has believed to have 124.43 million domestic ruminants; 60.39 million cattle, 31.30 million sheep and 32.74 goats, which was the largest population within the region of Africa. While livestock has historically played a significant role in the country’s economy, the value derived from these animals has been limited due to a variety of factors. Livestock diseases are one of the major roadblocks to the sector’s development, as they reduce productivity and impede commerce in animals and animal products [1,2]. Lumpy skin disease is one of the most common and widespread livestock diseases in all of the country’s regions [3].
A lumpy skin disease, caused by the lumpy skin disease virus, is a serious pox disease that affects cattle and Asian water buffalo (LSDV). It is among of the most economically significant viral diseases identified by the OIE as notifiable transboundary animal diseases, and the second most significant cattle disease in Ethiopia [4,5]. It is because of its economic significance that the disease listed on the OIE’s list of notifiable terrestrial animal diseases [6] and causes significant economic losses by reducing milk production, emaciation and poor growth in infected animals, permanent damage to hides, abortion, temporary or permanent infertility, and secondary bacterial infections can sometimes result in death [7,8].
LSDV is found under the genus Capripox virus (CaPVs) in the sub-family Chordopoxvirinae of Poxviridae family member. The Poxviridae family is distinguished by its huge and complicated genome, which consists a single, linear molecule of ds DNA that codes for approximately 200 proteins and consists of two subfamilies: Chordopoxvirinae, the vertebrate poxvirus, and Entomopoxvirinae, an insect poxvirus. The genus Capripoxvirus comprises: Lumpy skin disease virus (LSDV) and pox virus of sheep and goat (SPPV and GTPV) [9].
LSDV has limited host range and hence, can complete its replication cycle only in ruminant hosts. The disease mainly affects cattle and cattle of all ages, both sexes and all breeds are affected and is more severe in lactating and pregnant cows [8,10,11]. There is some evidence, however, that young animals are more vulnerable to the severe form of the disease; Bos indicus is less susceptible to clinical disease than Bos taurus, and Asian water buffaloes also have been reported to be susceptible. Despite the fact that LSD has not been reported in goats and sheep, characteristic skin lesions in sheep, goats, giraffes, impalas, and Grant’s gazelles kept in close contact with infected cattle have been established without systemic disease [12-14].
The entry of cattle from the affected region, as well as high temperatures and humidity, are usually linked to an LSDV outbreak in a previously disease-free area [7,15]. The virus must be fully disseminated to sensitive cattle in the nearby farms or surrounds for an outbreak to start and emerge after the initial exposure of sick animals into a new region [16]. It’s more common during the wet summer and autumn months, particularly in low-lying areas or around pools of water, but outbreaks can occur at any time of year [17]. The most major sources of infection to animals are thought to be; blood, nasal discharge, lacrimal secretions, semen and saliva. Mechanical vectors for the disease include blood-feeding insects like mosquitoes and flies [15].
In endemic area, vaccination is the only economically accepted way to control the spread of LSD and improve cattle productivity, because avoiding animal movement and affected animals removal alone are usually not effective [16,17]. LSDV and SPPV attenuated strains are used as vaccine strains in infected areas in the control of LSD. I case of vaccination, there is possibility of mild or systemic post-vaccination reactions in vaccinated animals. So, the application of diagnostic procedures that will rapidly and specifically differentiate LSDV field strains from LSD vaccine virus strains are extremely important [6].
Because of the disease’s widespread prevalence in Ethiopia, exporting live cattle and their products is a major challenge. Furthermore, the decline in milk and meat production, as well as the low quality of skin and hides, has a detrimental problem on national economic growth [5]. Since the country has no a welldesigned control strategy for this disease it is continuing to be a great problem. Even if the animal health authorities undertake vaccination campaigns when outbreak is reported, researches have shown that the different vaccines used in Ethiopia are not fully effective [18,19]. In addition, the lack of genetic information on in-field circulating virus and their association to the vaccine in use which is important for better matching of vaccines is also a major problem in the country [5]. Therefore, the objective of the study was to isolate and detect the virus responsible for the LSD occurrence in Yayo district using molecular techniques.
Materials and Methods
Description of the study area
The study was carried out in two selected Peasant associations (PA’s) (Shono and Bacho) of Yayo districts located approximately at 564 kilometers West to Addis Ababa in Illubabor zone of Oromia Regional State, Ethiopia (Figure 1). Yayo is located at latitude of 8o 26` 8`` N and longitude of 36o 20` 97`` E. The average rainfall Yayo is 1,200-2,800 mm which extends from February to November in normal years. The district has a land area of approximately 84,626 hectares and an elevation of 1400-2010 meters above sea level. The annual minimum and maximum temperature of Yayo is 18oC and 27oC. The domestic animals reared in Yayo district are 60,202 cattle, 38,386 poultry, 9,925 equines, 30120 sheep, and 35,120 goats. According to Yayo Woreda Agricultural Office, both livestock rearing and crop production are the main source of income of the majority of communities (about 87%) [2,20].
Figure 1: Map showing the study area.
Study animals (Populations)
Animals in the study were extensively managed, local breed cattle of 24 households of the Shono and Bacho PAs of Yayo district. All ages and both sexes of animals having suspected clinical signs of LSD were included in the study.
Study design
Outbreak investigation: An active disease investigation was carried out based on the reports of the LSD outbreak to NAHDIC through animal health professionals working in zonal animal health offices, and district animal health centers during the study period. The diagnosis of LSD depends on the basis of the signs and symptoms of affected cattle’s.
Sampling method and sample collection
In the present study, two PA’s (Shono and Bacho) were selected based on the reports of the outbreak case information gathered. Based on clinical examination of susceptible cattle’s (355 cattle’s), about 55 cattle’s were affected from which 44 cattle’s were selected and sampled from 24 small householders. The animals were selected after looking for clear signs and symptoms of lumpy skin disease. Animals that were critically sick were used to collect samples for viral isolation and molecular detection [12].
The samples were collected purposively from sick cattle. Aseptic skin scraping was performed with a scalpel blood after removing hair, washing and cleaning the area. Universal bottle containing phosphate buffer saline (PBS) with 2% antibiotic-antimycotic solution, prepared as virus transport medium at a pH of 7.2-7.6 [19,20] were used to contain tissue samples, which were kept in ice box and transported to molecular and virology laboratory of NAHDIC for laboratory test.
Laboratory techniques
Viral isolation: The biopsy samples removed from deep freezer were kept at room temperature and washed three times with sterile PBS with 7.2 approximate pH. About 1gm biopsy sample were triturated with mortar and pestle using sterile sand and mixed with 9ml sterile PBS containing antibiotic and antimycotic solution to have 10% suspension. The tissue suspension was then centrifuged at 3000rpm for 10min and the supernatant was filtered through a syringe filter of 0.45μm pore size (Millipore, United States of America (USA)). Approximately 0.1ml of the samples was inoculated in to sub-confluent VERO cell cultures in six wells plate along with cell controls. After 60min adsorption at 37oC, maintenance medium was added to each well including a negative control and incubated at 37oC in a humidified incubator at 5% CO2. Cells were monitored every 24hrs post-infection and inspected for cytopathic effects (CPEs) using an inverted microscope. On 6th day, the cultures were freezethawed and the resulting lysates were again inoculated into fresh cultures until the third passage [12,21].
DNA extraction: DNA was extracted from tissue samples using QIAmp viral DNA mini kit (Qiagen) according to the manufacturer’s protocol. First, 20μl of proteinase K was added to all tubes according to the sample size, then 200μl collected supernatants was added and 200μl AL buffer was added and mixed together by vortex mixer and incubated in a water bath at 56°C for 10 minutes and centrifuged briefly. To bind the nucleic acid on a mini spin column, 200μl of ethanol (100%) was added and mixed thoroughly with the help of vortex mixer for 15 seconds and briefly centrifuged. The mixture was transferred in to DNeasy minispin column in 2ml collection tube and centrifuged at 8,000rpm for 1min. The spin column having the DNA was transferred in to new 2ml collection tube and the first washing buffer 500μl AW1 added and centrifuged at 8000rpm for 1min. 500μl of second washing buffer, which is AW2, was added and centrifuged at 14000 rpm for 3 minutes and the filtrate was discarded. This step was repeated for 1 minute without adding any buffer. Then the mini spin column transferred to micro centrifuge tube and 200μl of AE elution buffer added and incubated at room temperature for 1-5 minutes to increase the yield of DNA and eluted by centrifugation at 8000rpm for 1 minute using fifth edition QiAamp DNA extraction protocol from Qiagen, 2016.
Conventional polymerase chain reaction (C-PCR)
A polymerase chain reaction was performed to detect the virus using Capripox virus specific primer: Forward and reverse having the sequence (SPPVDIV-F) 5′ATCTGCTACAAGTTTTAACGAACTTA 3′ and (SPPVDIV-R) 5′ TGAATGTGATCTCATATCCTTATTG-3′, respectively. DNA amplification was run in a final reaction volume of 20μl, containing 2μl of each forward and reverse primers and 2μl of 0.2mM dNTPs, 0.25μl Taq DNA polymerase (QIAGEN), 2μl of 10 x PCR buffer, 9.75μl of Rnase free H2O and 2μl template DNA. All PCR was performed using amplification program: 95°C for 4min followed by 35 cycles of 95°C for 30sec, 58°C for 30sec, and 72°C for 30sec and a final extension at 72°C for 2min to complete the amplification process [22].
To confirm the presence of DNA on the gel band, the amplified result was subjected to agarose gel electrophoresis [22]. The PCR products are separated by electrophoresis, using 2% agarose gel containing DNA staining dye (gel red) for 1hr at 100V and subsequently visualized under UV light. When 338bp were obtained, which is the anticipated amplification size for the LSDV genomic region, the results were considered positive for LSDV DNA.
Real time polymerase chain reaction (RT-PCR)
The virus was also detected using an RT-PCR test with Forward and Reverse Capripoxvirus specific primers having the sequence (Cap-B22RDIV-F) 5′TATGGATTTAGGAGTAGA3′ and (Cap- B22RDIV-F) 5′GCTTTACTTTAATATCATTG 3′, respectively. DNA amplification was carried out in a final volume of 20μl reaction mixture consisting of 10μl EVA green master, 2μl of each primer (Forward and Reverse) (4μl), 4μl of Rnase free H2O and 2μl of the sample DNA. The polymerase chain reaction was run using the following amplification program; initial denaturation at 95°C for 4min, followed by 42 cycles of 95°C for 5sec, 58°C for 5sec and 72°C for 5sec. The PCR products denatured at 95°C for 30sec, cooled to 65°C for 1min, and melted from 65°C to 90°C for 10sec for melting curve analysis. In each set of reactions, positives control plasmids and negative controls consisting of nuclease-free water in place of the template DNA were included. The positive samples were identified using cycle threshold (Ct) values from the test, which were utilized to describe the positive samples following amplification of the DNA template: The tissue specimens were considered negative when there is no or higher than 40 Ct value were obtained.
Data management and analysis
All field data was put into a Microsoft Excel spreadsheet and then uploaded to the Statistical Package for Social Sciences Software (SPSS) versions 20. The percentage of mortality calculated as number of death x100 over number of animals at risk and the fatality rate calculated as number of death x100 over number of sick animals. In addition, Chi-square (x²) test was used to assess the association of sex and ages with affected group. In both cases of the analysis, the confidence interval was set at 95% and P <0.05 was considered statistically significant [23].
Results
Outbreak investigation
From the total of 355 animals clinically examined, 55 cattle were affected and 5 died. The disease affected both sexes and all age groups of cattle. Lumpy skin disease outbreaks on cattle had experienced with morbidity and mortality during the study period. The observed morbidity, mortality and case fatality rate was 15.73%, 1.52% and 9.67% consequently in Shono kebele and 15.18%, 1.26% and 8.33% consequently in Bacho kebele (Table 1).
District
PA
Host
Number of animals at risk
Morbidity rate in %
Number of deaths
Mortality rate in %
Case fatality rate in %
Yayo
Shono
Cattle
197
31(15.73)
3
1.52
9.67
Bacho
158
24(15.18)
2
1.26
8.33
Total
355
55(15.49)
5
1.4
9.09
Abbreviation: PA: Peasant Associations.
Table 1: Morbidity, mortality and case fatality rates with respect to PAs of LSD outbreak.
Morbidity, mortality and case fatality rates were also assessed within age groups of <2 and ≥2 years old cattle’s and female and male cattle’s (Table 2 and 3).
Variables
Number of cattle at risk
Number of cattle affected
Morbidity rate in %
X2
P-value
Age
<2
99
11
11.11
2.215
0.137
≥2
256
44
17.18
Total
355
55
15.49
Sex
M
163
23
14.11
0.44
0.507
F
192
32
16.66
Total
355
55
15.49
Abbreviation: X2, Chi-square.
Table 2: Morbidity rate with respect to age and sex.
Variables
Number of cattle at risk
Number of cattle affected
Number of death
Mortality rate (%)
Case fatality rate (%)
Age
<2
99
11
1
1.01
9.09
≥2
256
44
4
1.56
9.09
Sex
M
163
23
3
1.84
13.04
F
192
32
2
1.04
6.25
Total
355
55
5
1.4
9.09
Table 3: Mortality and case fatality rates of LSD according to age and sex.
Viral isolation
Out of all (44) samples inoculated on vero cell line, characteristic CPE of capripoxvirus was observed on 17 samples following postinoculation after two or three blind passages whereas, virus isolates could not be identified from the remaining five skin samples. The characteristic CPE observed were rounding of single cells, aggregation of dead cells and destruction of monolayer. None of the negative control cultures showed any CPE after two or three blind passages (Figure 2).
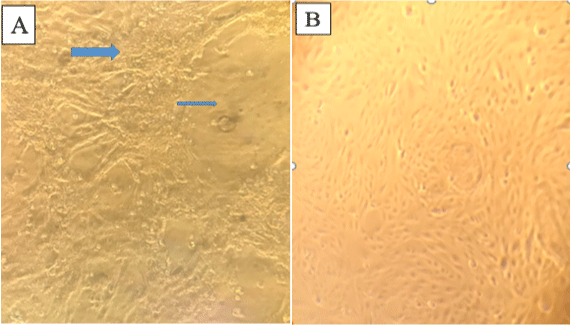
Figure 2: Photo of Vero cell: A) CPE positive on Vero cell after 3 passage post inoculation (The arrow indicates the aggregative and destruction of cell monolayers);
B) Normal cell monolayer.
Detection of lumpy skin disease virus by conventional PCR
The 44 extracted DNA samples were amplified using forward and reverse capripoxvirus specific primers having the sequence SPPVDIV: 5′ATCTGCTACA AGTTTTAACGAACTTA 3′ and SPPVDIV: 5′TGAATGTGATCTCATATCCTTATTG 3′ respectively from which 88.63% (39/44) were detected by conventional PCR. The molecular weight of the amplified PCR product was 338bp (Figure 3), which is the expected size for the targeted genomic region of LSDV. The resulting LSDV PCR products are uniformly aligned on line, indicating that they have the same sample amplicons size. The amplified PCR product had a molecular weight of 338bp, which is the anticipated size for the LSDV genomic region (Figure 3). The positive LSDV PCR products are consistently matched on the line, showing the same sample amplification size.
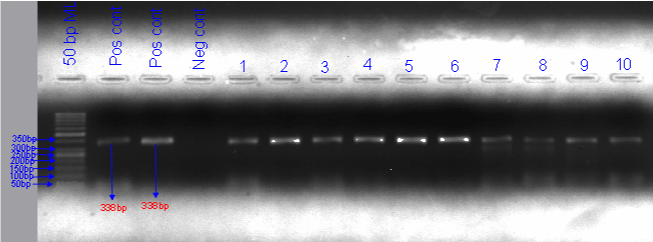
Figure 3: Conventional PCR based detection of LSDV in samples taken from skin nodules of infected animals.
ML: Molecular Ladder; Pos cnt: Positive Control; Neg cnt: Negative Control.
Detection of lumpy skin disease virus real time PCR
Out of 44 extracted DNA samples amplified by real time PCR, 42 samples (95%) of them were found positive. The samples and control real time PCR amplification curve was indicated below (Figure 4).
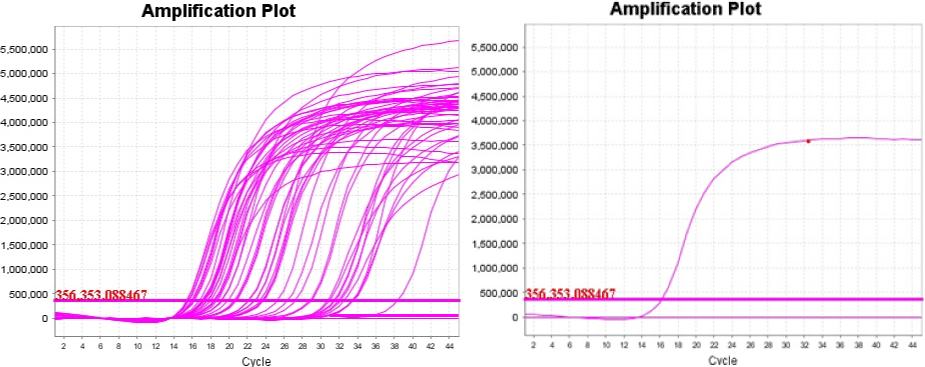
Figure 4: Real time PCR amplified samples (left) and control (right).
As shown in the table below (Table 4), positive samples have Ct values ranging from 15.38-32.19. Some of the values are lower when compared to the positive LSD control Ct values (16.01 and 16.19), indicating the high virus concentrations. Undet displayed the negative specimens, showing undetermined values or very high Ct values (around 41).
Kebele
Sample No
Ct value
Kebele
Sample No
Ct value
Positive control
16.01
Shono
S23
20.72
Shono
S1
17.29
Shono
S24
27.23
Shono
S2
15.42
Bacho
S25
20.55
Shono
S3
18.66
Bacho
S26
20.72
Shono
S4
15.66
Bacho
S27
20.8
Shono
S5
17.33
Bacho
S28
22.13
Shono
S6
16.81
Bacho
S29
22.18
Shono
S7
15.57
Bacho
S30
22.76
Shono
S8
16.15
Bacho
S31
22.78
Shono
S9
15.98
Bacho
S32
23.31
Shono
S10
18.35
Bacho
S33
23.55
Shono
S11
17.25
Bacho
S34
23.65
Shono
S12
22.78
Bacho
S35
23.72
Shono
S13
22.13
Bacho
S36
25.25
Shono
S14
27.25
Bacho
S37
26.41
Shono
S15
18.35
Bacho
S38
27.25
Shono
S16
23.55
Bacho
S39
28.77
Shono
S17
23.31
Bacho
S40
28.92
Shono
S18
19.2
Bacho
S41
31.58
Shono
S19
Undet.
Bacho
S42
32.19
Shono
S20
23.65
Bacho
S43
33.43
Shono
S21
28.77
Bacho
S44
Undet.
Shono
S22
17.98
Negative control
Undet.
Note: Undet indicate undetermined or very high Ct values (around 41), S indicates sample.
Table 4: Real time PCR Ct values of samples collected from Shono and Bacho PAs of Yayo district.
Discussion
Lumpy skin disease outbreaks from two peasant associations (PA) of Yayo district were investigated in the present study. The occurrence of the disease was examined and confirmed using clinical diagnosis, virus isolation and PCR. Fever, skin nodules, swollen lymph nodes, lameness, depression, lacrimation, and salivation were the most and common clinical characteristics of LSD observed during these outbreaks. Other authors have also reported the same symptoms in natural and experimental infections [24-26].
The overall morbidity, mortality and case fatality rates of the present study were 15.49%, 1.4% and 9.09% respectively, indicating the high economic effect of the disease in the area. The morbidity rate (15.49%) observed were closer to the report in central Ethiopia with 13.61% [21] and reported 6.1% [27], which is slightly lower than the current finding. There is also wide range of morbidity rates (3% up to 85%) reported by other authors [7]. The present finding of mortality rate (1.4%) and case fatality rate (9.09%) of LSDV in cattle was lower than [21] who reported 4.97% mortality rate and 36.49% case fatality rate and [27] who reported 1.8% and 30% mortality and case fatality rate respectively. Differences in climate and geographic location, management conditions, immune status and condition of the animals, virus pathogenicity, and the type and quantity of insect vectors could all contribute to these discrepancies [7].
Although the difference was not statistically significant (P>0.05), higher morbidity rate was observed in female (16.66%) than male (14.11%). In contrast, mortality and case fatality rate were higher in male (1.84 and 13.04%, respectively) than female (1.04 and 6.25%, respectively). Despite the fact that oxen are used to plough the land and thus may be stressed, they are rarely exposed to the disease since they are kept separate from the herd around the house, keeping them away from infected animals and the vector’s replication area. The morbidity rates may have risen in female because of physiological (like pregnancy and lactation) and management conditions. Female animals are generally kept together and managed extensively, which could facilitate the transmission of the disease in between the animals. While Shono (15.73, 1.52 and 9.67%) and Bacho (15.18, 1.26 and 8.33%) PAs have the same morbidity, mortality and case fatality rates of LSD, respectively. This might be due to the similarity of agroecological condition the two PAs. Both areas are closer to rivers which might be suitable for the replication of arthropod vectors.
The highest morbidity (17.18%) was observed in adult than young (11.1%). This is inline with [28] report but it disagrees with Previous data that mentioned calves were more susceptible to LSD infection than adult cattle [21,29]. This disparity may be due to the disease’s occurrence in the herd, which increased the numbers of infected cattle ≥2 years (adult) and the management conditions of adult animals, specially females cattle extensive management might compromise their immunity.
Lumpy skin disease (SDV) can be cultured in a number of caprine, ovine, or bovine primary cells or cell lines. The LSDV has been adapted to grow on the chorioallantoic membrane of embryonated chicken eggs and African green monkey kidney (VERO) cells [12]. It develops slowly in cell cultures, and CPE is usually detectable five to seven days after inoculation [30]. A typical CPE can be detected, which includes retraction of the cell membrane from surrounding cells, rounding of individual cells, and nuclear chromatin margination [12]. In this outbreak investigation the virus was isolated by growing on VERO cells (P-38) and CPE characterized by aggregated cells, destruction of cell monolayers and rounding of cells were observed on 3rd passage 5th day. Similar CPE characteristics were recorded by other authors [21].
Many notable scientific breakthroughs have resulted from the invention of traditional PCR and real-time PCR. Though both methods are still widely used, real-time PCR is gaining greater popularity as the most cost- and time-effective approach for evaluating DNA products. Both techniques were used for identification of the virus responsible for the outbreak. Conventional PCR test used in this study revealed a unique band with the anticipate size of 338bp. Real time PCR Ct values taken from the positive samples indicate lower numbers lying around the Ct values of positive controls. Undetermined values or very high Ct values (around 41) were indicated as negatives in which lower or no loads of the virus are present.
Conclusion and Recommendations
The present study showed that, LSDV is circulating in cattle in the Yayo district of Illubabor zone. Regardless of the difference in sex and age, the disease affected both sexes and all age groups of cattle and has already caused significant economic loss due to reduced production, damage to hides, and death. The occurrence of LSD in the study area was confirmed by cell culture and real time and conventional PCR that the outbreaks were due to LSDV. So, Active LSDV search, detecting and characterization of the virus should be continued and further study on control strategy is necessary. To reduce economic losses caused by the disease, strategic programs for effective control and prevention should be established.
Declaration
Availability of data and materials: The data sets used during the current study was available from the corresponding author on reasonable request.
Ethical approval: Ethical clearance obtained from the Animal Research Scientific and Ethics Review Committee of the National Animal Health Diagnostic and Investigation Center (NAHDIC). Before conducting this research, all the animal owners were informed about the purpose of the study and also, they are given well aware of the importance and benefit of the research in terms of immediate and future values. Safe handling procedures were followed while collecting samples. For notification, formal letters were written and sent to the study district and animal owners by the National animal health diagnostics and investigation center to reach on consent.
Acknowledgments: Our sincere thanks go to National animal health diagnostics and investigation center (NAHDIC), for financial support during the study period and Mr. Abebe Olani for assisting us in technical works. We are exceptionally grateful to Dr. Esayas Gelaye, Vice director of National Veterinary Institute for allowing us to use the laboratory reagent. We are also greatly indebted to Illubabor zone and Yayo district animal health office heads and veterinarians as well as Shono and Bacho Peasant association (PA) clinic veterinarians for sincerely helping us out in the whole study period and the animal owners who without them we couldn’t have accomplish this work.
Author contributions: All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
References
- Mekonnen S, Hussein I, Bedane B. The distribution of ixodid ticks (Acari: Ixodidae) in central Ethiopia. Onderstepoort J. Vet. Res. 2001; 68: 243-251.
- CSA. Federal Democratic Republic of Ethiopia Central Statistical Agency Agricultural Sample Survey Volume II Report on Livestock and Livestock Characteristics (Private Peasant Holdings). 2018.
- Teshome D. Prevalence of major skin diseases in ruminants and its associated risk factors at University of Gondar Veterinary Clinic, North West Ethiopia. J Res Dev. 2016; 4: 1-7.
- Gari G, Bonnet P, Roger F, Waret-Szkuta A. Epidemiological aspects and financial impact of lumpy skin disease in Ethiopia. Prev Vet Med. 2011; 102: 274-283.
- Gelaye E, Belay A, Ayelet G, Jenberie S, Yami M, Loitsch A, et al. Capripox disease in Ethiopia: Genetic differences between field isolates and vaccine strain, and implications for vaccination failure. Antivir. Res. 2015; 119: 28-35.
- Vidanović D, Šek*ler M, Petrović T, Debeljak Z, Vasković N, Matović K, et al. Real-time PCR assays for the specific detection of field Balkan strains of lumpy skin disease virus. Acta Vet. 2016; 66: 444-454.
- Tuppurainen ES, Oura CA. lumpy skin disease: an emerging threat to Europe, the Middle East and Asia. Transbound Emerg Dis. 2012; 59: 40-48.
- Gumbe AA. Review on lumpy skin disease and its economic impacts in Ethiopia. J. Dairy Vet. Anim. Res. 2018; 7: 39-46.
- Tulman ER, Afonso CL, Lu Z, Zsak L, Kutish GF, Rock DL. Genome of lumpy skin disease virus. J. Med. Virol. 2001; 75: 7122-7130.
- Abdallah FM, El Damaty HM, Kotb GF. Sporadic cases of lumpy skin disease among cattle in Sharkia province, Egypt: Genetic characterization of lumpy skin disease virus isolates and pathological findings. Vet. World. 2018; 11: 1150.
- Mammo B. Isolation, Molecular Characterization and Vaccine Effectiveness Study of Lumpy Skin Disease Virus in Selected Diary Farms of Central Ethiopia. J. Biol. Agricult. H. Care. 2019; 9: 1-13.
- OIE. Terristerial manual chapter 2.4.13. Lumpy Skin Disease. 2017.
- Carn VM, Kitching RP. The clinical response of cattle experimentally infected with lumpy skin disease (Neethling) virus. Arch virol. 1995; 140: 503-513.
- Davies FG. Lumpy skin disease of cattle: a growing problem in Africa and the Near East. World Animal Review. 1991; 68: 37-42.
- Tulman ER, Afonso CL, Lu Z, Zsak L, Sur JH, Sandybaev NT, et al. The genomes of sheeppox and goatpox viruses. J. Med. Virol. 2002; 76: 6054- 6061.
- Sprygin A, Pestova Y, Wallace DB, Tuppurainen E, Kononov AV. Transmission of lumpy skin disease virus: A short review. Virus Res. 2019; 269: 197637.
- Nir O, Braverman Y, Davidson M, Grinstein H, Haymovitch M, Zamir O. Spread of lumpy skin disease in Israeli dairy herds. Veterinary Record. 1995; 137: 91-93.
- Ayelet G, Abate Y, Sisay T, Nigussie H, Gelaye E, Jemberie S, et al. Lumpy skin disease: preliminary vaccine efficacy assessment and overview on outbreak impact in dairy cattle at Debre Zeit, central Ethiopia. Antivir. Res. 2013; 98: 261-265.
- Molla W, de Jong MC, Frankena K. Temporal and spatial distribution of lumpy skin disease outbreaks in Ethiopia in the period 2000 to 2015. BMC Vet Res. 2017; 13: 1-9.
- YWAO. Yayo wereda agricultural office socio economic data. 2019.
- Ayelet G, Haftu R, Jemberie S, Belay A, Gelaye E, Sibhat B, et al. Lumpy skin disease in cattle in central Ethiopia: outbreak investigation and isolation and molecular detection of the virus. Rev. Sci. Tech. 2014; 33: 877-887.
- Chibssa TR, Grabherr R, Loitsch A, Settypalli TB, Tuppurainen E, Nwankpa N, et al. A gel-based PCR method to differentiate sheeppox virus field isolates from vaccine strains. J. Med. Virol. 2018; 15: 1-7.
- Moulton LH, Wolff MC, Brenneman G, Santosham M. Case-cohort analysis of case-coverage studies of vaccine effectiveness. Am. J. Epidemiol. 1995; 142: 1000-1006.
- Agag BI, Mousa S, Hassan HB, Saber MS, El-Deghidy NS, El-Aziz AM. Clinical, serological and biochemical studies on lumpy skin disease. J. Appl. Anim. Res. 1992; 1: 13-23.
- Body M, Singh KP, Hussain MH, Al-Rawahi A, Al-Maawali M, Al-Lamki K, et al. Clinico-histopathological findings and PCR based diagnosis of lumpy skin disease in the Sultanate of Oman. Pak. Vet. J. 2012; 32: 206-210.
- Jalali SM, Rasooli A, Seifi Abad Shapuri M, Daneshi M. Clinical, hematologic, and biochemical findings in cattle infected with lumpy skin disease during an outbreak in southwest Iran. Arch. Razi Inst. 2017; 72: 255-265.
- Alemayehu G, Leta S, Eshetu E, Mandefro A. Incidence of lumpy skin disease and associated risk factors among export-oriented cattle feedlots at Adama District, Central Ethiopia. J. Vet. Med. Anim. Health. 2015; 7: 128-134.
- Kasem S, Saleh M, Qasim I, Hashim O, Alkarar A, Abu-Obeida A, et al. Outbreak investigation and molecular diagnosis of Lumpy skin disease among livestock in Saudi Arabia 2016. Transbound Emerg Dis. 2018; 65: e494-500.
- Ahmed WM, Zaher KS. Observations on lumpy skin disease in local Egyptian cows with emphasis on its impact on ovarian function. Afr. J. Microbiol. Res. 2008; 2: 252-257.
- Tuppurainen E. Diagnostic assays for the detection of lumpy skin disease virus and antibodies. EMPRES. 2017; 47: 7-9.