
Research Article
Austin J Vet Sci & Anim Husb. 2022; 9(3): 1098.
Isolation and Antibiogram of Escherichia coli Isolated from Selected Dairy Farm at Sebeta, Oromia, Ethiopia
Shubisa A²*, Sintayehu S¹ and Mekonnen A¹
¹Animal Health Institute (AHI), Sebeta, P.O. Box, 04, Ethiopia
²Jimma University College of Agriculture and Veterinary Medicine, Ethiopia
*Corresponding author: Shubisa Abera Leliso, Animal Health Institute (AHI), Ethiopia
Received: August 08, 2022; Accepted: August 26, 2022; Published: September 02, 2022
Abstract
Mastitis is the most prevalent disease of dairy animals, imparting huge economic losses to the dairy industry. Escherichia coli (E. coli) bacteria are among the common causes of mastitis in dairy animals. A cross-sectional study was carried out from March 2021 to August 2021 on lactating Cattle suffering from mastitis cases to isolate Escherichia coli and to assess their antimicrobial susceptibility pattern in Sebeta town, Oromia Special Zones, Ethiopia. Prevalence of mastitis by California Mastitis Test at cow level was found to be 87.14% (122/140), out of which 12.85% (18/140) and 74.28% (104/140) were clinical and subclinical, respectively. The study revealed that out of the 560 quarters examined, 24 (4.2%) of them were blind and 536 were functional. The Escherichia coli isolated from CMT positive was 8.07% (31/384); from this clinically and sub clinically affected udder were 1.04% (4/384) and 7.03% (27/384) respectively. Finally, the antimicrobial profiles of the 31 E. coli confirmed isolates were assessed using 10 different antimicrobials. Out of the 31 isolates tested, 7 (22.58%) isolates were found to be highly resistant to Ampicillin, 5 (16.12%) isolates were resistant to Trimethoprim+Sulphamethoxazole, and 3 (9.67%) and 4(12.9%) isolates were developed resistant to Amoxicillin+Cluvanate acid and Tetracycline respectively. However, all thirty-one isolates of Escherichia coli were found to be highly susceptible to five antimicrobials namely Ciprofloxacin (100%), Cefoxitin (100%), Meropenem (100%), Cefotaxime (100%) and Gentamycin (100%). In conclusion, this study determined the importance of Escherichia coli as one of mastitis causing bacterium of the dairy industry and investigated its antimicrobial resistance pattern. Thus, researchers would like to emphasize the need for urgent intervention to control the diseases and prevent the associated loss.
Keywords; Antimicrobial resistance; CMT; Sebeta; Mastitis; Escherichia coli
Introduction
Mastitis is an inflammation of the mammary glands. It causes a great loss or reduction in animal productivity than any disease of dairy cattle. It is the costliest disease and remains a series problem for dairy industry for its influences both on the quantity and quality of milk produced it causes a marked fall in milk yield and is a cause of culling of animals [1].
It is the disease that cause burden in dairy livestock all over the world, the persistency of the microbes leads to invasion of tissue that can opportunistically results in mastitis. Due to invasion and bacterial infection mammary glands release white blood cells leading to secretion of toxins in response to immune response that trigger mastitis infection in bovine. There are several bacteria involved in causing mastitis however most infections are caused by Gram-negative rods like Escherichia coli or Gram-positive cocci mainly Staphylococcus and streptococci depending on the mode of transmission and host condition [2].
Escherichia coli are abacterium commonly found in gastrointestinal tract of animals and humans (Welch 2006). Most of the Escherichia coli are non-pathogenic or harmless [3] but some of them can be pathogenic if they are opportunistic [4] and commonly affect those individuals of immune-challenged. Pathogenic Escherichia coli are usually the types that can cause diarrhea when transmitted through contaminated water and foods [5].
E. coli frequently contaminates food organism and it is a good indicator of fecal pollution [6]. Presence of E. coli in milk products indicates the presence of enteropathogenic microorganisms, which constitute a public health hazard. E. coli is among many pathogenic microorganisms which can access to milk and some of dairy products which considered a reliable indicator of contamination by manure, soil and contaminated water [7]. In Ethiopia, the consumption of raw milk is very traditional and contaminated milk and milk products are the most common transmission pathway of E. coli from animals to humans. Even though the disease caused by E. coli is very important in the country and of great public health concern, E. coli has received very little consideration in many of the previous public health studies. Most of the previous studies circulated in limited areas and fail to represent the incidence of E. coli under different management and ecological situations.
The antibiotics are routinely practiced as therapeutic measures of this malady but indiscreet use of antibiotics develops resistance in animal body which lowers cure rate. That all can lead to low therapy success and increased risk to human health due to uncontrolled use of antibiotics [8]. The development of resistance of the bacteria to antimicrobial agents makes mastitis more difficult to control. Antimicrobial resistance has become a huge public health issue worldwide [9]. Moreover, in the country both veterinary and medical drugs are often misused, creating ideal conditions for t he development of resistant strains, thus better understanding of the antimicrobial susceptibility/resistance/patterns of pathogens isolated from animal source foods like milk is needed. There was no recent study with regard to the prevalence of mastitis, isolation of E. coli and antibiogram pattern of the isolates among dairy cattle at Sebeta town. Therefore, this study was conducted with the following objectives
• To isolate and determine antibiogram profile of E. coli isolates from selected dairy farms in the study area.
Materials and Methods
Study Area
The study was conducted in Sebeta town. Sebeta is an urban set up located about 26 kilometers southwestern of Addis Ababa (Figure 1). Its geographical location is 08°9200 North and 38°6200 east. The mean annual rainfall and temperature of the town are 1073 milliliters and 17.40°C, respectively. The altitude ranges from 2356-2405 m above sea level. The conurbation was purposely selected mainly due to presence of many commercial and semi-commercial farms.
Figure 1: Map of the Study Area.
Study Design
A cross sectional study was conducted to generate the desired data from March 2021 to August 2021.
Sample Size Determination
The required sample size of this study was determined by the formula given by [10] based on the 4.3% mean expected previous prevalence [11]; 95% confidence interval and 5% desired precision. The sample size was calculated as follows.
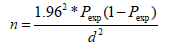
Where, N: required sample size; Pexp: expected prevalence, and d²; desired absolute precision of 0.05. Therefore, the calculated sample size was 63, but to increase the precision of the study and to increase the number of E.coli isolates, from a total of 140 dairy cows, 384 milk samples was collected from dairy farms in Sebeta Town.
Study Population
The study was carried out on 140 lactating exotic, local and crossbreed dairy cows kept under intensive, semi-intensive and extensive farming system in the study area. Nineteen dairy farms located in Sebeta Town were included in the study. Then simple random sampling was employed to select individual cows. The study involved physical examination of sampled cows and collection of milk samples following standard procedures. The samples from each functional udder of cows were screened for sub-clinical mastitis using the California Mastitis Test (CMT).
Milk Sample Collection and Transportation
During milk sample collection, cows were restrained in standing position and sampling began with teat cleaning by scrubbing thoroughly using cotton balls moistened with 70% alcohol. Milk samples were collected first from the closest teats followed by those at the far side of the udder by maintaining universal bottle at approximately 45° angles. The fore strip milk was discarded and 10- 15 ml of midstream milk sample was taken. The universal bottles were labeled for information such as date of collection, name of farm and cow identification number [12]. Milk Samples were transported using cold chain (Ice box) from dairy farms to Animal Health Institute (AHI) located in Sebeta Town.
Sample Storage and Processing
Most mastitis causing microorganisms survive under refrigeration for several days or freezing for several weeks. The milk samples were stored in the laboratory at +4°C until laboratory examination is conducted. CMT positive milk samples were inoculated on media and incubated at 37°C for 24–48 hours.
Bacterial Isolation and Identification
Bacterial isolation and identification were conducted using the guideline prescribed by [13]. Since the primary and main step of any bacteriological isolation starts with preparation of media, the study started by preparing different selective, differential and indicator media that were needed for the research. Brain Heart Infusion Broth, Brain Heart Infusion agar, MacConkey agar, Eosin Methylene Blue (EMB) agar, Nutrient agar, and Muller-Hinton agar media were prepared whenever they were needed to do colony characterization and to perform further biochemical, confirmatory and susceptibility tests.Secondary biochemical test of E. coli isolates was performed using gram staining, IMViC and Triple Sugar Iron (TSI) slant agar test.
MALDI-TOF Biotyper Identification System
Final confirmation was done by MALDI-TOF, according to the manufacturer’s guidelines [14]. A single young colony of sub cultured bacteria was directly deposited on a MALDI-TOF plate (Bruker Daltonik GmbH, Karlsruhe, Germany). At the end of sample 0.5 μl BTS were added to the plate. The sample was mixed with 0.5 μl of matrix solution and placed on the steel surface of the target plate to dry. The matrix solution (cinnamic acid or a benzoic acid derivate) co-crystallizes with the sample on the target plate. The loaded target plate is inserted into the machine where it was then transported to the measuring chamber. Within the mass spectrometer, a high vacuum has to be continuously maintained. However, upon insertion of the loaded target plate, air is introduced into the system and the vacuum must be reestablished before sample analysis can be performed. Once a sufficient vacuum has been created, the individual samples were exposed to short laser pulses. The laser’s energy vaporizes the microorganism together with the matrix, leading to ionization of the (ribosomal) proteins. An electromagnetic field, created by a potential of about 20 kV, accelerates the ions before they enter the flight tube. The Time of Flight (TOF) of the analytes to reach the detector at the end of the flight tube was precisely measured. The degree of ionization as well as the mass of the proteins determines their individual TOF. Based on this TOF information, a characteristic spectrum was recorded and constitutes a specific sample fingerprint, which was unique for a given species. This virtual gel view represented all of the peaks in a spectral file and was used to compare the spectra of the E. coli isolates that were tested. Clusters with similar protein expression were identified by the Principal Component Analysis (PCA) [15]. Overall, thirty-one isolates were confirmed as Escherichiacoli by MALDI-TOF tests and were preserved in a brain heart infusion broth with glycerol broth to conduct antimicrobial susceptibility tests.
Antimicrobial Susceptibility Test
Disc Diffusion Susceptibility Test: Antimicrobial Susceptibility test of the isolates Escherichia coli was analyzed for ten different antimicrobials namely Amoxicillin and Cluvanate acid (30μg), Tetracycline (30μg), Meropenem (10μg), Ciprofloxacin (10μg), Ceftriaxone (30μg), Cefoxitin (30μg), Sulphamethoxazole+Trimethoprim (25μg), Ampicillin (10μg), Cefotaxime (30μg) and Gentamycin (10μg). The antimicrobials were selected based on their availability at the laboratories.
The test was carried out by using the agar disc diffusion method, first 5ml of 0.85% saline water was dispensed in a test tube labeled for 31 isolates and the colonies from a brain heart infusion agar media was then taken by a disposable plastic loop and put in the saline water and each suspension with the organism was mixed well and measured for a turbidity of 0.5 McFarland standards and cultured thoroughly on Muller- Hinton Agar (MHA) media of 4mm depth and 90mm diameter. After that the ten different antimicrobial discs listed above were taken from their corresponding containers by using forceps and diffused in the respective MHA Media of the thirty-one isolates and incubated at 370C for 18 hours. After incubation, measurement of the diameter of the clear Zones of Inhibition (ZOIs) around and including the antimicrobial discs of each isolate were conducted and interpreted to categorize as susceptible, intermediate and resistant according to the performance standards given by Clinical and Laboratory Standards Institute [16].
Data Analysis
Descriptive statistics such as frequency, percentage, and/or proportion were used for prevalence, antimicrobial resistance test. Chi-squire test (χ²) was used to assess significant differences by Statistical Package for Social Science (SPSS) of version 20 software. The results with less than P-value of 0.05 were considered statistically significant.
Results
California Mastitis Test
The prevalence of mastitis in this study was found to be 87.14% (122/140) using CMT. From this 12.85% (18/140) were clinical and 74.28% (104/140) were subclinical cases. Out of the 560 quarters 24 of them were blind (4.2%) and 536 were functional. From a total of 536 quarters, 33 of them were showing clinical and 503 were subclinical forms of mastitis. Hence the overall prevalence of mastitis in our study was confirmed to be 384. From a total of 536 quarters tested for mastitis by CMT Test, 6.15% (33/536) were clinical and 65.48% (351/536) subclinical (Table 1).
Farm
Blind quarter
Negative quarter
Positive quarter
Total Examined
Prevalence %
1
1
8
19
28
71.42%
2
0
12
16
28
57.14%
3
3
17
28
48
64.58%
4
1
15
40
56
73.21%
5
4
16
48
68
76.47%
6
0
12
20
32
62.50%
7
0
8
16
24
66.66%
8
3
13
24
40
67.50%
9
1
11
8
20
45.00%
10
2
6
16
24
75.00%
11
2
2
24
28
92.85%
12
0
4
28
32
87.50%
13
0
0
12
12
100%
14
1
3
16
20
85.00%
15
2
10
20
32
68.75%
16
2
6
12
20
70.00%
17
0
2
18
20
90.00%
18
1
5
10
16
68.75%
19
1
2
9
12
83.33%
Total
24
152
384
560
73%
Table 1: Prevalence of mastitis at quarter level in selected dairy farms of Sebeta Town.
The above table indicates that overall prevalence at quarter level found to be 73% in selected dairy farm in Sebeta town.
Isolation and Identification of E. coli
The results of the present study revealed that out of 384 CMT positive milk samples, 31 samples were found to be positive for E. coli. Isolates were characterized as bright pink color on MacConkey agar plates and showed blue-greenish metallic sheen on EMB agar plate. Upon Gram’s staining of the isolates under 100x using light microscope, pink-colored, small rod-shaped organisms arranged in single, pairs or short-chain were identified. The biochemical characteristics of 31 E. coli isolate showed positive for catalase, Methyl red and Indole test but negative for Voges-Proskauer, Urease, and Citrate. In addition, reactions in TSI agar slant revealed yellow but with gas and production of hydrogen sulfide was observed. Almost all the isolates of E. coli fermented lactose, sucrose and glucose with the production of both acid and gas. Based on the results from the biochemical test further identification was done MALDI-TOF to confirm the bacteria.
MALDI -TOF Identification
The 31 isolates that were identified as E.coli by biochemical test were again subjected to MALDI-TOF Biotyper for confirmation. A total of 31 (8.07%) isolates were confirmed as Escherichia coli by MALDI-TOF identification system (Table 2 and 3). Out of these 1.04% clinical and 7.03% were subclinical. The E. coli isolated at cow level was 19.67% and out of this 12.12% were clinical and 7.55% were subclinical. All isolates were analyzed for 10 different antimicrobials to assess their antibiotic profiles and categorize them as susceptible, intermediate or resistant.
Sample Name
Sample ID
Organism (best match)
Score Value
Organism (second-best match)
Score Value
A1 (+++) (A)
19214 (Standard)
Escherichia coli
2.29
Escherichia coli
2.29
A2 (+++) (A)
17856 (Standard)
Escherichia coli
2.38
Escherichia coli
2.33
A3 (+++) (A)
17867 (Standard)
Escherichia coli
2.18
Escherichia coli
2.15
A4 (+++) (A)
19188 (Standard)
Escherichia coli
2.38
Escherichia coli
2.37
A5 (+++) (A)
19184 (Standard)
Escherichia coli
2.40
Escherichia coli
2.37
A6 (+++) (A)
19200 (Standard)
Escherichia coli
2.41
Escherichia coli
2.31
A7 (+++) (A)
19191 (Standard)
Escherichia coli
2.33
Escherichia coli
2.30
Result overview table--continued on next page
Table 2: A table shows the similarity of Organism best match and Organism second best match.
Samples tested
Rank (Quality)
Matched Pattern
Score Value
NCBI Identified
1
(+++)
Escherichia coli DSM 1578 DSM
2.29
562
2
(+++)
Escherichia coli DSM 682 DSM
2.38
562
3
(+++)
Escherichia coli DH5alpha BRL
2.18
562
4
(+++)
Escherichia coli RV412 A1 2010 06a LBK
2.38
562
5
(+++)
Escherichia coli BM11464 1 CHB
2.40
562
6
(+++)
Escherichia coli ATCC 25922 CHB
2.41
562
7
(+++)
Escherichia coli ESBL EA 1528T CHB
2.33
562
8
(+++)
Escherichia coli ATCC 35218 CHB
2.24
562
9
(+++)
Escherichia coli DSM 1103 QC DSM
2.29
562
10
(+++)
Escherichia coli ESBL 1528T CHB
2.35
562
11
(+++)
Escherichia coli ATCC 25922 THL
2.29
562
12
(+++)
Escherichia coli Nissl VML
2.36
562
13
(+++)
Escherichia coli ATCC 25922 THL
2.38
562
14
(+++)
Escherichia coli DH5alpha BRL
2.34
562
15
(+++)
Escherichia coliDH5alpha BRL
2.37
562
16
(+++)
Escherichia coli DSM 682 DSM
2.32
562
17
(+++)
Escherichia coli RV412 A1 2010 06a LBK
2.27
562
18
(+++)
Escherichia coli MB 11464 1
2.38
562
19
(+++)
Escherichia coliDSM 1576 DSM
2.31
562
20
(+++)
Escherichia coli RV412 A1 2010 06a LBK
2.28
562
21
(+++)
Escherichia coli DSM 1576 DSM
2.34
562
22
(+++)
Escherichia coli DH5alpha BRL
2.22
562
23
(+++)
Escherichia coli DH5alpha BRL
2.33
562
24
(+++)
Escherichia coli DH5alpha BRL
2.25
562
25
(+++)
Escherichia coli DSM 682 DSM
2.35
562
26
(+++)
Escherichia coli DSM 1576 DSM
2.33
562
27
(+++)
Escherichia coli DSM 1576 DSM
2.16
562
28
(+++)
Escherichia coli MB 11464 1
2.21
562
29
(+++)
Escherichia coli DSM 1576 DSM
2.23
562
30
(+++)
Escherichia coli RV412 A1 2010 06a LBK
2.32
562
31
(+++)
Escherichia coli DSM 1576 DSM
2.31
562
Table 3: A table shows rank, matched pattern and score value of E. coli.
Antimicrobial Resistance Test
A total of 31 isolates were subjected to the 10 antibacterial agents for AMR test. The AMR test results showed that E. coli is 100% susceptible to six antimicrobial agents and showed different percentage of resistance to five antimicrobials (Table 4).
Antimicrobial agents
Potency
Interpretation of results (zone diameter in mm)
µg/disk
S (%)
I (%)
R (%)
Ciprofloxacin
5μg
31(100%)
0 (0%)
0 (0%)
Cefotaxime
30μg
31(100%)
0 (0%)
0 (0%)
Cefoxitin
30μg
28(90.32%)
1 (3.2%)
2(6.45%)
Amoxicillin + cluvanate acid
30μg
27(87.1%)
1 (3.2%)
3 (9.6%)
Trimethoprim + Sulphamethoxazole
25μg
26(83.87%)
0 (0%)
5(16.12%)
Meropenem
10μg
31(100%)
0 (0%)
0 (0%)
Gentamycin
10μg
31 (100%)
0 (0%)
0 (0%)
Ampicillin
10μg
22(70.96%)
2(6.45%)
7(22.58%)
Tetracycline
30μg
27(87.09%)
0 (0%)
4 (12.9%)
Ceftriaxone
30μg
31(100%)
0 (0%)
0 (0%)
%=Percent, S=Sensitive, I=Intermediate, R=Resistant,
Table 4: Antibiogram Pattern of E. coli isolates to 10 different antimicrobials.
As shown in Table 3, all 31 isolates of Escherichia coli were found to be highly susceptible to five antimicrobials namely Ciprofloxacin (100%), Meropenem (100%), Ceftriaxone (100%), Cefotaxime (100%) and Gentamicin (100%). However, 7 isolates were also found to be highly resistant to Ampicillin (22.58%) followed by 5 isolates resistant to Trimethoprim+sulphamethoxazole (16.12%) and also 4, 3 and 2 isolates were found to be resistant to Tetracycline (12.9%), Amoxicillin+clavulanate acid (9.67%) and Cefoxitin 2(6.45%) respectively. The isolates were also indicating an intermediate resistance for Amoxicillin + clavulanate acid,Cefoxitin 1(3.2% for each) and Ampicillin 2(6.45%) (Table 4). Two isolates were resistance to more than two antibiotics.
Risk Analysis
In this study, the occurrence of E. coli was significantly associated with different parity numbers (χ²=7.7706; P=0.021) (Table 5). The highest percentages of E. coli isolates were isolated from cows with eight and above parity number 2 (20%) and from poor body condition 8 (12.30%). On the other hand, the present finding revealed that the association between different groups of age, lactation stage, breed, body condition and quarter with the occurrence of E. coli organisms were not statistically significant (x²=2.4077; P= 0.300; (x²=2.4012; P= 0.301);x2=0.7449; P= 0.689; χ²=1.9610; P= 0.375; χ²= 0.6859; P= 0.877 respectively (Table 5).
Variables
No. of samples examined
No. of positive (%)
Chi-square(χ2)
P-Value
Body condition
Good
242
18 (7.34%)
1.9610
0.375
Medium
77
5 (6.49%)
Poor
65
8 (12.30%)
Age
Young
27
2 (7.40%)
2.4077
0.300
Adult
332
29 (8.73%)
Breed
Exotic
325
26 (8%)
0.7449
0.689
Cross
22
1(4.54%)
Local
37
4 (10.81%)
Parity
Few
185
8 (4.32%)
7.7706
0.021
Moderate
189
21 (11.11%)
Many
10
2 (20%)
Quarter
RH
94
9 (9.57%)
0.6859
0.877
RF
95
6 (6.31%)
LH
98
8 (8.16%)
LF
97
8 (8.24%)
Lactation
Early
21
1 (4.76%)
2.4012
0.301
Medium
276
26 (9.42%)
Late
87
4 (4.59%)
Key: RH= Right Hind; RF= Right Front; LH= Left Hind and LF= Left Front
Table 5: Prevalence of E. coli occurrence with host and environment related factors.
As the above table indicates, the occurrence of mastitis by E. coli were higher in poor body condition of animal (12.30%) when compared to those medium and very good body condition (6.49%) and (7.34%) respectively. Based on the parity of the animal, the highest percentages of E. coli isolates were isolated from cows with eight and above parity number (20%) when compared to those of parity number greater than or equal to three and less than or equal to seven (moderate) (11.11%)and less than three parity number (few) (4.32%). A high prevalence of E. coli was detected in right hind (9.57%) and left-fore quarter (8.24%) followed by left hind (8.16%) and right-fore quarter (6.31%).
Discussion
In our study the CMT test result showed the prevalence of mastitis in to be 87.14% (122/140). Our finding was similar to [17] who observed 86.2% cases of mastitis through CMT screening of in dairy cows in the study conducted at Kampala, Uganda in 2014 but it was lower in comparison with finding of [18], and [19] who reported that the overall mastitis prevalence in the farm was 66.6% and 71.0% in Assella Dairy farm and Holeta town respectively. This finding was not in agreement with those of [20] and [21] who reported the prevalence of 52.78% in Ethiopia and 52% in Nigeria, respectively.
In the current study, a total of 384 raw milk samples taken from 560 quarters were examined bacteriologically and biochemical tests were performed to detect Escherichia coli. All E. coli isolates were able to produce bright pink-colored colonies on MacConkey agar, characteristic metallic sheen colonies on the EMB agar.
The overall E. coli isolated among mastitis positive cattle in this study 19.67% was in agreement with the finding of [22] who reported 18.6% prevalence of E. coli in the study conducted at Benchi Maji Zone, Southwest Ethiopia in 2015.Similarly, our result was in agreement with [23] who reported 20.9% prevalence of E. coli in raw milk in Iran in 2021. However, our finding was lower than the finding of [24] who reported 40.7% prevalence of E.coli among dairy cows in Pakistan in 2004. Our finding was higher than the previous reports of [25] at Holeta (4.6%) and [19] in and around Sebeta (0.75%). The prevalence of E. coli is probably due to the fact that E. coli is the commonest environmental contaminants which are closely associated with hygiene. It becomes pathogenic whenever the hygienic conditions of the animal or environment become poor. In addition, the existence of high concentration of E. coli in milk also indicates the relatively poor quality of milk, related with substandard hygiene of the farm management.
Out of the total E.coli isolates (19.67%) in this study, the specific occurrence of E.coli in clinical and sub clinical cases was known to be 12.12% and 7.55%respectively. This finding was in agreement with [26,27] who reported 11.5 and 3.64% prevalence of E.coli from clinical and sub clinical cases respectively. The current finding was lower when compared to previous reports of [28,29] who observed 34.9 and 17.04% respectively. E. coli is a poor contagious bacterium, and the highest prevalence of other pathogens of mastitis particularly Staphylococcus, Streptococcus and Corynebacterium lead to rise in somatic cell count of milk which limits the multiplication of the coliforms has also shown that Pseudomonas is intermittently shed from the udder and does not frequently appear in milk.
In our study from 384 fresh raw milk samples, 8.07% milk samples were known to be contaminated with E. coli. This was in agreement with the studies by [30-32] who reported 8.75%, 10.3% and 11.2% from Malaysia, Ghana and Hawassa in Southern Ethiopia respectively. The current finding was lower as compared to [33] who found 21% from India and [34] who reported quarter prevalence of 17.9%. The recent study was relatively higher as compared to the report by [35] who found 3.88% prevalence of E. coli from raw milk of cows in Iraq. The differences in the prevalence of E. coli in our study and other researchers could be attributed to variations in the study area, sample size, methodology we used and season.
In this study, the prevalence of clinical and subclinical mastitis in the study area was 1.04% and 7.03% respectively. This was in agreement with [36] who reported the prevalence of clinical and subclinical mastitis 8.3% and 1.8% respectively, lactating cows from smallholder dairy farms in Sellalle area, Central Ethiopia. The difference in the recent study from past studies might be attributed to differences in environmental conditions, management differences in the production system (intensive, semi-intensive, and extensive), ecology, hygienic practices and methodological differences among these studies.
In the present study, the occurrence of E. coli has been found significantly associated with different parity numbers of (x²=7.7706; P=0.021) which is in agreement with reported by [37,20,38,39]. On the other hand, the present finding revealed that there was no association between different groups of age, lactation stage, breed and body condition with the occurrence of E. coli and our results agree with that of [32] in cows in Hawassa, who reported no association of the prevalence of mastitis with age, lactation stage, body condition and history of mastitis.
In this study the highest percentages of E. coli isolates were isolated from cows with eight and above parity number (20%) when compared to those of parity number greater than or equal to three and less than or equal to seven (moderate) (11.11%) and less than three parity number (few) (4.32%). This finding was in agreement with [40] who reported 31.5% from cows with age group from seven to ten years. This could be due to multiple parturition stresses and this ultimately down regulates their immunity, and immunity normally decreases as the animal gets older making more prone to E. coli infection [40].
A high prevalence of E. coli was detected in right hind (9.57%) and left-fore quarter (8.24%) followed by left hind (8.16%) and rightfore quarter (6.31%). This finding is in agreement with that of [25] who reported a high prevalence in hind quarter, in contrast, [41,42] reported a high prevalence in four quarters. The high prevalence in hind quarters might presumably be associated with increased chance of hind quarters being soiled with urine and faces or by the tail leading to poor udder management [43].
A total of 31 E. coli isolates were tested against 10 antimicrobials based on CLSI guidelines and all E. coli isolates were found to be 100% susceptible to gentamicin Ciprofloxacin, Menoperem, Ceftriaxone and Cefotaxime followed by Tetracycline (87.1%), Amoxicillin + cluvanate acid (87.09%), sulphamethoxazole-trimethoprim (83.87%) and Ampicillin (70.96%). All isolated E. coli were found to be 22.58% resistant to Ampicillin followed by Trimethoprim + sulphamethoxazole (16.12%), Tetracycline (12.9%), Amoxicillin + clavulanate acid (9.67%) and Cefoxitin (6.45%). Relatively similar findings have been reported by [44] All E. coli isolates were 100% susceptible to gentamicin, 15% resistant to sulphamethoxazoletrimethoprim from Burkinafaso. The result of this study was almost comparable with the work of [45,46] who reported the susceptibility against E. coli were 100% for Ciprofloxacin and Gentamycin and resistance against Tetracycline (86.88%from Ethiopia and Pakistan respectively. Our result was relatively less than [47] who reported sulphamethoxazole-trimthoprim (76%) was susceptible to E. coli from Mekelle, Ethiopia. Therefore, in this study gentamycin, Ciprofloxacin, Meropenem, Ceftriaxone and Cefotaxime were found to be the most effective drugs against E. coli infection in the study area.
Conclusion and Recommendations
• This study clearly indicates that fresh raw cow’s milk was found to be highly contaminated with the E. coli organisms. Since many people still drink fresh raw milk without further heat processing, it is a serious public health problem as milk is a vehicle for food borne diseases. Risk factors like parity number, has significant association with occurrence of E. coli; whereas there was no significant difference among different age, breed, lactation stage and body condition. Based on the antimicrobial susceptibility pattern, E. coli isolates were found to be highly susceptible to gentamicin, ciprofloxacin, meropenem, ceftriaxone and cefotaxime whereas resistant to ampicillin, trimethoprim+sulphamethoxazole, tetracycline, amoxicillin and clavulanate acid. Based on the above remarks, the following recommendations need to be considered:
• To ensure the quality of raw milk, everyone engaged in milk and dairy production chain should be trained for hygienic practices.
• In order to protect consumers from zoonotic AMR, food safety management programs should be implemented and highly considered.
• Awareness should be given to the community at risk, whole sellers and distributers.
• Consistent teat dips should be applied after milking.
Declarations Ethics Approval and Consent to Participate
Animal Health Institute (AHI) Research Ethics Review Committee (WUREC) approved this research before actual data collection. A consent sheet was prepared in English and attached to the tool on a separate page regarding the purpose, description, anticipated benefits, and other relevant aspects of the study, and signed informed consent was taken from all respondents prior to data collection for animal owners of above 18 years of age. Thus, the authors declare that all methods were performed in accordance with the relevant guidelines and regulations.
Availability of Materials and Data
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to the manuscript to the final submission. Conceptualization, Data curation, analysis, and writing the original draft were performed by Shubisa Abera, Investigation, methodology, validation, and supervision were majorly done by Mekonnen Addis while visualization, reviewing, and editing was done by Shubisa Abera. Finally, all authors read and approved the final manuscript submission.
Aknowledgements
Above all thanks goes to GOD for his innumerable favors and fruit full results. I do have also special spiritual respect for his mother saint merry. Next, I would like to express my deepest.
Gratitude and heartfelt thanks to my advisor Dr. Mekonnen Addis and co-advisor from AHI Dr. Shubisa Abera, General bacteriology staff Abebe Olani, Mekdes Tesfaye, Letebrhan Yemesgen for the guidance, encouragement, valuable suggestions and constructive comments.
My sincere gratitude also goes to Animal Health Institute and Jimma University College of Agriculture and veterinary medicine for their indispensable encouragement, financial and moral support throughout my academic career.
Lastly, my grateful appreciation also extends to my family for their endless love and support throughout my academic career. The cooperation and support by the owners of dairy cows during data collection and examination of animals are highly acknowledged.
References
- Blum S, Heller ED, Krifucks O, Sela S, Hammer-Muntz O, Leitner G. Identification of a bovine mastitis Escherichia coli subset. Veterinary microbiology. 2008; 132: 135-148.
- Abebe R, Hatiya H, Abera M, Megersa B, Asmare K. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Veterinary Research. 2016; 12.
- CDC. Raw Milk Questions and Answers. 2017.
- Akond M, Akond MA, Alam S, Hassan SM. R, Shirin M. Antibiotic Resistance of Escherichia coli isolated From Poultry and Poultry Environment of Bangladesh. Internet Journal of Food Safety. 2009; 11: 19-23.
- Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clinical Microbiology Reviews. 2013; 26: 822-880.
- Benkerroum N, Bouhlal Y, Attar AE, Marhaben A. Occurrence of Shiga toxin-producing Escherichia coli O157 in selected dairy and meat products marketed in the city of Rabat, Morocco. Journal of food protection. 2004; 67: 1234-1237.
- Soomro A, Arain M, Khaskheli M. and Bhutto B. Isolation of Escherichia coli from raw milk and milk products in relation to public health sold under market condition at Tandojam. Pakistan Journal of Nutrition. 2002; 13: 151-152.
- Barkema HW, Schukken YH, Zadoks RN. Invited Review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. Journal of dairy science. 2006; 89: 1877- 1895.
- Anueyiagu KN, Isiyaku AW. Isolation, identification of Staphylococcus aureus from bovine milk and its antibiotics susceptibility. International Journal of Livestock Production. 2015; 6: 74-77.
- Thrusfield M. Veterinary epidemiology. 3rd edition London: Blackwell Science. Oxfored, UK. 2007; 227-247.
- Dadi S, Lakew M, Seid M. Koran T. and Olani A. Isolation of Salmonella and E. coli (E. coli O157:H7) and its Antimicrobial Resistance Pat- tern from Bulk Tank Raw Milk in Sebeta Town, Ethiopia. J Anim Res Vet Sci. 2016; 4: 021.
- Ott SL, Costs of herd-level production losses associated with subclinical mastitis in U.S. dairy cows. Proceedings of the 38th Annual Meeting on National Mastitis Council, Feb. 14-17, USA, Pp: 2001; 152-152.
- Quinnn M, A, Conaghan G, Greenstein A, Karim Z, Brown C. Emery P. Sustained response in early poor prognosis RA after withdrawal of infliximab therapy. In Arthritis and Rheumatism. 2002; 46: 3416-3416.
- Van Veen SQ, Claas EJ, Kuijper EJ. High-Throughput Identification of Bacteria and Yeast by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in Conventional Medical Microbiology Laboratories. J Clin Microbiol. 2010; 48: 900–907.
- Egli A, Tschudin-Sutter S, Oberle M, Goldenberger D, Frei R, Widmer AF. Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass- Spectrometry (MALDI-TOF MS) Based Typing of Extended-Spectrum β-Lactamase Producing E. coli – A Novel Tool for Real-Time Outbreak Investigation. PLoS ONE. 2015; 10: e0120624.
- CLSI Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and laboratory standards institute. 2020.
- Abrahmsén M, Persson Y, Kanyima BM, Båge R. Prevalence of subclinical mastitis in dairy farms in urban and peri-urban areas of Kampala, Uganda. Tropical Animal Health and Production. 2013; 46: 99-105.
- Abera B, Lemma D, Iticha I. Study of bovine mastitis in Asella government dairy farm of Oromia Regional state, South Eastern Ethiopia. International journal of current research and academic review. 2013; 1: 134-145.
- Hundera S, Adema Z, Sintayhue A. Dairy cattle mastitis in and around sebeta, Ethiopia. Int J Appl Res Vet Med. 2005; 3: 332-338.
- Mekonnen SA, Koop G, Melkie ST, Getahun CD, Hogeveen H, Lam TJGM. Prevalence of subclinical mastitis and associated risk factors at cow and herd level in dairy farms in North-West Ethiopia. Preventive veterinary medicine. 2017; 145: 23-31.
- Salih MD, Junnaid AU, Tambulual FM, Magaji AA. Tafaku S. Prevalence of bovine mastitis in lactating cow in same selected commercial dairy farms in Sokota Metroplis. Appl Sci Res. 2011; 2: 290- 294.
- Biruke D, Shimeles A. Isolation and identification of major bacterial pathogen from clinical mastitis cow raw milk in Addis Ababa, Ethiopia. Academic Journal of Animal Diseases. 2015; 4: 44-51.
- Momtaz H, Farzan R, Rahimi E, Dehkordi FS, Souod N. Molecular Characterization of Shiga Toxin-Producing Escherichia coli Isolated from Ruminant and Donkey Raw Milk Samples and Traditional Dairy Products in Iran. The Scientific World Journal. 2012; 2012: 1-13.
- Iqbal M, Khan MA, Daraz B and Saddique U. Bacteriology of mastitic milk and in vitro antibiogram of the isolates. Pakistan Veterinary Journal. 2004; 24: 161-164.
- Mekibib B, Furgasa M, Abunna F, Megersa B, Regassa A. Bovine mastitis: prevalence, risk factors and major pathogens in dairy farms of Holeta town, Central Ethiopia. Veterinary World. 2010; 3: 397.
- Zerihun T. A study on Bovine Sub-Clinical Mastitis at Stela Dairy Farm, DVM Thesis, FVM, A.A.U. Ethiopia. 2006.
- Bayush T, Abera A. Prevalence of Mastitis and Associated risk factors in Jimma Town Dairy Farms, Western Ethiopia. J Vet Sci Ani Husb. 2018; 6: 304.
- Tola T. Discussion, Bovine Mastitis in indigenous Arsi Breed and Arsi Hlesteinrosses. DVM Thesis, FVM, A.A.U. Ethiopia. 2006.
- Haile T. Discussion: Bovine Mastitis in Indigenous zebu and Boran-Holestein crosses in South Wollo, DVM thesis, FVM, AAU, Ethiopia. 2005.
- Lye L, Afsah LH, Chang S, Loo Y. Puspanadan S. Risk of Escherichia coli O157:H7 transmission linked to the consumption of raw milk. International Food Research Journal; 2013; 20: 1001-1005.
- Addo K, Mensah G, Aning K, Nartey N, Nipah G. Risk of Escherichia coli O157:H7 transmission linked to the consumption of raw milk in the state of Ghana. International Food Research Journal. 2011; 20: 1001-1005.
- Abera M, Habte T, Aragaw K, Asmare K, Sheferaw D. Major causes of mastitis and associated risk factors in smallholder dairy farms in and around Hawassa, Southern Ethiopia. Tropical Animal Health and Production. 2011; 44: 1175-1179.
- Mohanty N, Das P, Pany S, Sarangi N, Ranabijuli S. Isolation and antibiogram of E. coli isolates from clinical and subclinical cases of bovine mastitis. Veterinary World. 2013; 6: 739-743.
- Almaw G. Cross sectional study of bovine mastitis in and around Bahirdar and antibiotic resistance patterns on major pathogens. MSc thesis, Addis Ababa University, Faculty of Veterinary medicine, Ethiopia. 2004: 79.
- Madhloom IH. Drawing of the evolutionary tree for mastitis infections caused by E. coli in Basrah province. (Master Thesis), College of Veterinary Medicine, University of Basrah. 2017.
- Getahun K, Kelay B, Bekana M, Lobago F. Bovine mastitis and antibiotic resistance patterns in Selalle smallholder dairy farms, central Ethiopia. Tropical Animal Health and Production. 2007; 40: 261-268.
- Lakew BT, Fayera T, Ali YM. Risk factors for bovine mastitis with the isolation and identification of Streptococcus agalactiae from farms in and around Haramaya district, eastern Ethiopia. Tropical Animal Health and Production. 2019; 51: 1507-1513.
- Birhanu M, Leta S, Mamo G, Tesfaye S. Prevalence of bovine subclinical mastitis and isolation of its major causes in Bishoftu Town, Ethiopia. BMC Research Notes. 2017; 10.
- Sarba EJ, Tola GK. Cross-sectional study on bovine mastitis and its associated risk factors in Ambo district of West Shewa zone, Oromia, Ethiopia. Veterinary World. 2017; 10: 398-402.
- Rezaei M, Karimi F, Yahyaei M, Javdani H, Shahabi A. A survey of microbial total count and prevalence of Escherichia coli in raw milk in Markazi Province, Iran. Research Opinions in Animal and Veterinary Sciences. 2013; 12: 474- 477.
- Khanal T, Pandit A. Assessment of sub-clinical mastitis and its associated risk factors in dairy livestock of Lamjung, Nepal. International Journal of Infection and Microbiology. 2013; 2: 49-54.
- Kavitha L, K Rajesh K, Suresh K, Satheesh Syamasundar N. Buffalo mastitis risk factors. Buffalo bulletin. 2009; 28: 134-168.
- Hogan J, Smith KL. Coliform mastitis. Veterinary research. 2003; 34: 507- 519.
- Bagré TS, Kagambèga A, Bawa Ibrahim H, Bsadjo Tchamba G, Dembélé R, Zongo C, et al. Antibiotic susceptibility of Escherichia coli and Salmonella strains isolated from raw and curds milk consumed in Ouagadougou and Ziniaré, Burkina Faso. Afr. J. Microbiol. Res. 2014; 8:1012-1016.
- Bedasa S, Shiferaw D, Abraha A, Moges T. Occurrence and antimicrobial susceptibility profile of Escherichia coli O157:H7 from food of animal origin in Bishoftu town, Central Ethiopia. International Journal of Food Contamination. 2018; 5: 1-8.
- Momtaz H, Farzan R, Rahimi E, Safarpoor F, Dehkordi M, and Souod N. Molecular characterization of Shiga toxin-pro- ducing Escherichia coli isolated from ruminant and donkey raw milk samples and traditional dairy products in Iran, The Scientific World Journal. 2012; 2012: 231342.
- Yohannes G. Isolation, identification and antimicrobial susceptibility testing of Escherichia coli isolated from selected dairy farms in and around Mekelle, Ethiopia. J Dairy Vet Anim Re. 2018; 7: 287‒291.