Research Article
Austin Virol and Retrovirology. 2014;1(1): 5.
Identification of Pseudocowpox Virus in Angus Bull with Failure to Breed
Wendy Black1, Michael T Walburger3, Rocky Baker1, Claire Ostertag-Hill1, Aimee Reed2, Margo Whipple1 and Ling Jin1,2*
1Department of Biomedical Sciences, Oregon State University, US
2Department of Microbiology, Oregon State University, US
3Rocky Mountain Large Animal Clinic, US
*Corresponding author: Ling Jin, Department of Biomedical Sciences, College of Veterinary Medicine, Oregon State University, Corvallis, OR 97331, USA
Received: July 25, 2014; Accepted: August 19, 2014; Published: August 21, 2014
Abstract
Pseudocowpox, a Parapoxvirus, causes a common mild infection of the udder and teats of cows. Infection of genital organs has not been reported before in bulls. In this report, pseudocowpox virus (PCPV) was identified from one of two 1-year-old Angus bulls (Bos taurus) with red rashes and papule lesions on the surface of the penis. Inoculation of bovine turbinate cells or primary bovine testicular cells with the swab of the papule lesions produced CPE characteristic of pox virus infection. Parapoxvirus like particles were observed by EM in viral pellets of the tissue culture isolate. To confirm the virus, designated 14V10296 isolate, was a Parapoxvirus, viral DNA of the 14V10296 isolate was examined by PCR with pan Parapoxvirus primer sets that could amplify B2L gene of all four Parapoxviruses. The expected B2L amplicons of the 14V10296 isolate showed 99% homology to B2L of PCPV strain F00.120R. To further confirm the isolated virus is PCPV, PCR primers specific for Orf vascular endothelial growth factor gene and IL-10 and PCPV uracil DNA glycosidase (UDG) and IL-10, respectively, were used to amplify the viral DNA of the 14V10296 isolate. Only when PCR primers specific to PCPV UDG and IL-10 were used, expected products were amplified from viral DNA of the 14V10296 isolate, respectively. Phylogenetic analysis suggests the 14V10296 isolate is closely related to PCPV strain F00.120R, an isolate from reindeer. These results from tissue culture, EM, PCR amplification and DNA sequence analysis suggest that the bulls were infected by a PCPV which is closely related to PCPV strain F00.120R.
Keywords: Pseudocowpox; Parapoxvirus; Angus bull
Introduction
Pseudocowpox virus causes a common, mild infection of the udder and teats of cows. This virus is widespread worldwide. Pseudocowpox virus (PCPV) is a member of genus Parapoxvirus in the family Poxviridae, which includes bovine popular stomatitis virus (BPSV), contagious ecthyma virus (Orf) in sheep and goats, and Parapoxvirus of red deer [1]. These Parapoxviruses differ morphologically from other poxviruses of other genera. Lesions from Parapoxvirus infections normally begin as small, red papules on the teats or udder and are followed rapidly by scabbing or by the formation of small vesicles [2,3]. Infection of PCPV is self-limiting and normally resolves in about 2 weeks; however, some lesions may persist for several months, giving the affected teats a rough feel and appearance and more scabs may form. The infection spreads slowly throughout milking herds and a variable percentage of cows show lesions at any one time. Cattle may become re-infected in subsequent lactations. Infection of PCPV is mostly associated with the udder and teats [4]. Infection of genital tissue has not been reported in cattle. This study describes a PCPV, 14V10296 isolate that was isolated from a bull with failure to breed.
Materials and Methods
Case presentation
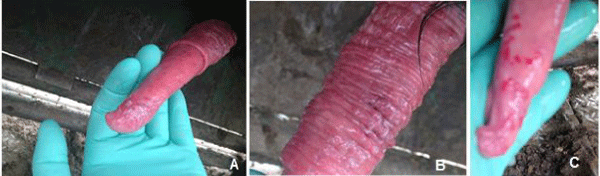
Two 1-year old Angus bulls were purchased from a pure bred breeder in Idaho. They were ranch raised, weaned and finished in allotments of 100-400 individuals. When exposed to cows, both bulls exhibited difficulty with intromission. They failed to breed in the spring of 2014 and were examined at Rocky Mountain Large Animal Clinic in Spanish Fork, UT. Poxvirus like lesions were seen on the prepuce and penis of bulls as red rashes (Figure 1C), papules, and vesicles (Figure 1A,1B). A preputial swab from one bull and preputial scraping from another bull were collected and sent to the Oregon State University Veterinary Diagnostic Lab for virus isolation.
Figure 1 : Pseudocowpox virus infected bull with lesions on the on the surface of the penis. A: Cutaneous lesions on the prepuce of bull A. B: Lesions and vesicles on the penis of bull A. C: Red rash lesion on the penis of bull B.
Tissue culture and virus isolation
Bovine Turbinate cells (BT cells) and primary bovine testicular cells (Testis cells), generated from newborn bovine testicles, were maintained in DMEM supplemented with 10 % gamma-irradiated fetal bovine serum (Atlas Biological Inc, CO). The sample preparations were filtered first in 0.45 μm syringe filter and then inoculated to 90% confluent monolayer of BT cells or Testis cells. Prior to absorption all cells were pretreated with 1X DEAE-Dextran (2.5 μg/ml). The inoculate was adsorbed on cells for one hour, then the cells were rinsed twice in PBS and maintained in DMEM supplemented with 5% gamma-irradiated fetal bovine serum (Atlas Biological Inc, CO), penicillin (100 U/ml) and streptomycin (100 μg/ml) (Sigma-Aldrich, Inc., St. Louis, MO) and incubated at 37°C.
Electron microscopy
The isolated virus was propagated in Testis cells maintained in DMEM supplemented with 5% fetal bovine serum and antibiotics as described above. Virus was harvested when more than 90% of the cells showed CPE. The infected flasks were subjected to one freeze– thaw cycle. The media harvested from infected Testis cells was cleared of cells and cell debris by centrifugation at 1500 rpm for 15 min in a clinical centrifuge. The virions were then centrifuged in a Beckman Model L8-70 ultracentrifuge at 29,000 rpm for 1 h in a 50.2 TI Rotor. Virus particles pelleted by ultracentrifugation were adsorbed on formvar-coated carbon-stabilized copper grids. Briefly, 10 μl of purified virus particles were mixed with 10 μl of 2% PTA (pH 6.9) in water, then a small drop was placed directly upon the form var coated carbon-stabilized copper grid. The grid was then blotted dry with Whatman filter paper and allowed to air dry. Images were obtained with a FEI Titan 80-200 TEM electron microscope.
Primers and PCR amplification
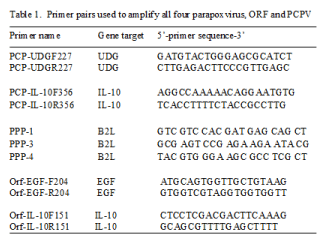
A set of primers, pan-parapoxvirus primer 1 (PPP-1) and PPP-4 were selected as reported previously [5], which is based on the major envelope protein (B2L) gene of Orf virus strain NZ2 (Accession No. DQ184476). A semi-nested primer PPP3 was also used as previously reported [5]. Primers Orf-EGF-F204 and Orf-EGF-R204 specific for Orf virus vascular endothelial growth factor gene (EGF) and primers Orf-IL-10F151 and Orf-IL-10R151 specific for Orf virus vIL-10 were selected based on Orf virus strain OV-SA00 (Accession No. AY386264). Primers PCP-UDGF227 and PCP-UDGR227 specific for PCPV uracil DNA glycosylase (UDG) and primers PCP-IL-10F356 and PCP-IL-10R356 specific for PCPV vIL-10 were based on PCPV DNA sequences available from NCBI (Accession No. JQ728421.1). Sequences of primers mentioned above are included in Table 1.
Table 1:
PCR amplification with specific primers for detection of viral DNA from tissue culture was performed as follows: a 25-μl solution consisting of 2.5 μl 10× amplification buffer (Lucigen), 0.5 μM MgSO4, 1.0 μl dNTPs at 10 mM each, 0.4 μM primers (forward and reverse), 1.0 U Econo Taq DNA polymerase (Lucigen), and 0.5–1 μg total tissue DNA. The mixture was subjected to 94°C for 2 min, and 30 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 45 s, followed by a 5-min elongation reaction at 72°C after the final cycle. Ten micro liters of each PCR sample was analyzed in a 1.5% agarose gel in TAE buffer (40 mM Tris-OH, 20 mM acetic acid, pH 7.8) and then visualized by UV illumination after staining with ethidium bromide (1 μg/ml). Commercially available 1 kb plus ladder (Life Technologies) served as size markers.
DNA sequencing
Sequences were determined by direct Sanger sequencing of PCR products after purification with a Charge Switch PCR Clean-Up kit (Invitrogen). All sequencing was carried out by the Center for Gene Research and Bio computing (CGRB) at Oregon State University. The CGRB used an ABI Prism®3730 Genetic Analyzer with a Big Dye® Terminator v. 3.1 Cycle Sequencing Kit, employing ABI Prism®3730 Data Collection Software v. 3.0 and ABI Prism® DNA Sequencing Analysis and Phylogenetic analysis of the DNA sequences were carried out using Geneious software.
Results
Virus Isolates
Following infection of the BT cells or Testis cells with the preputial lesion swab from one of the bulls, CPE was observed at 1-4 days post-infection and was characterized by enlarged cytoplasm, rounding, and cell detachment. The initial virus cultures were then passed again in BT cell and Testis cells. Similar CPE was observed in both BT and Testis cells following infection of the initial culture supernatant. The isolate of the virus was designated 14V10296 isolate. No virus was isolated from the scraping sample from the other bull.
Electron microscopy
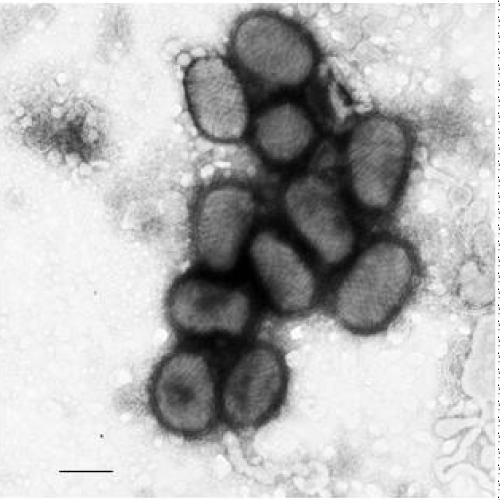
To determine the morphology of the virus, virions from the cell culture were pelleted by ultracentrifugation, subjected to negative staining and examined by transmission electron microscopy. As shown in Figure 2, virions from tissue culture have unique pine cone morphology. The viral particle has a long axis of about 260 nm on average and a short axis of about 150 nm. The surface of the virion has an array of tubule-like structures in a zigzag-cross manner on the surface of the virion. The morphology of the virions of the bull isolate is similar to other known Parapoxvirus virions [6,7].
Figure 2 : Electron micrographs of pseudocowpox virions isolated from lesion swab. Scale bar = 200 nm.
Viral DNA amplification with PCR
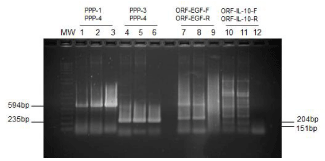
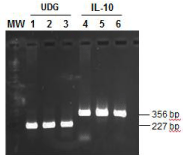
Viral DNA extracted from the infected tissue cultures was first examined by pan-parapoxvirus primers PPP-1 and PPP-4 or PPP-3 and PPP-4. As shown in Figure. 3, a 594 bp product and a 235 bp product, at the expected sizes, were amplified, respectively, from the total DNA extract of cells infected with second passage of the isolate (Figure 3, lane 1-2, 4-5) as well as with the first passage of the isolate (Figure 3 lanes 3 and 6) using both pairs of pan-parapoxvirus primers (Figure 3 lane 1-6). When primers specific for Orf virus -EGF or -IL-10 were used, no specific products were amplified (Figure 3 lane 7-12). When PCR primers specific for PCPV UDG or IL-10 were used, expected products were amplified from total DNA isolated from both passages (Figure 4). As shown in Figure 4, a 227 bp product was produced using primers PCP-UDGF227 and PCP-UDGR227 (Figure 4, lane 1-3), a 356 bp product was produced using PCP-IL10F356 and PCP-IL10R356 (Figure 4, lanes 4-6).
Figure 3 : Detection of PCPV DNA using PCR with pan-Parapoxvirus primers and orf specific primers. Lane 1-3: PCR amplification of viral DNA with primers PPP-1 and PPP-4; Lanes 4-6: PCR amplification of viral DNA with primers PPP-3 and PPP-4; Lanes 7-9: PCR amplification of viral DNA with primers Orf-EGF-F204 and Orf-EGF-R204 specific for Orf vascular endothelial growth factor gene (EGF); Lanes 10-12: PCR amplification of viral DNA with primers Orf-IL-10F151 and Orf-IL-10R151 specific for vIL-10. Lanes 1, 4, 7, and 10: 0.5 μg of total DNA extracted from tissue culture infected with the second passage of the 14V10296 isolate, Lanes 2, 5, 8, and 11: 1.0 μg of total DNA extracted from tissue culture infected with the second passage of the 14V10296 isolate. Lanes 3, 6, 9, and 12: 1.5 μg of total DNA extracted from tissue culture infected with the first passage of the 14V10296 isolate.
Figure 4 : Detection of PCPV DNA by PCR using PCPV specific primers. Lane 1-3: PCR amplification of viral DNA with primers PCP-UDGF227 and PCP-UDGR227 specific for PCPV-UDG; Lane 4-6: PCR amplification of viral DNA with primers PCP-IL-10F356 and PCP-IL-10R356 specific for PCPVvIL- 10. Lanes 1 and 4: 0.5 μg of Viral DNA extracted from tissue culture infected with the 14V10296 isolates, Lanes 2 and 5: 1.0 μg of Viral DNA extracted from tissue culture infected with the 14V10296 isolate. Lanes 3 and 6: Viral DNA extracted from lesion scrapes.
DNA sequence analysis and molecular phylogeny
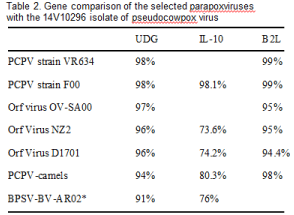
DNA sequence analysis of the PCR product amplified by primers PPP-1 and PPP-4 revealed that the sequence of 14V10296 B2L (530 bp) (Gen Bank accession No. KM277584) is 100%, 99% and 97-98% homologous to B2L DNA sequence of PCPV strain 00/03, PCPV F00. 120R and other PCPV major envelope protein (B2L) genes, respectively (Table 2). The B2L DNA sequence of the isolate has about 95% homology to Orf virus B2L gene (Gen Bank accession: KF837136) (Table 2).
Table 2:
To determine the relationship of the 14V10296 isolate to other members of the Parapoxvirus genus, the uracil DNA glycosidase (UDG) gene sequence amplified by PCR was sequenced and examined. A 181 bp sequence was obtained from the PCR product amplified from the total DNA extracts from cells infected with the first passage of 14V10296 isolate. As shown in Table 2, the UDG DNA sequence of the 14V10296 isolate has 98% homology to PCPV strain VR634 and F00.120R, 96% to Orf virus, 94% to PCPV from camels, and 91% to BPSV strain BV-AR02. Phylogenetically, the 14V10296 isolate is closer to strain F00.120R (Figure 5). It branched away from Orf virus-UDG similarly as PCPV-F00.120R. Although the DNA sequence of PCPV UDG was only 96% identical to Orf virus UDG, no amino acid coding difference in UDG protein was observed between PCPV and Orf virus.
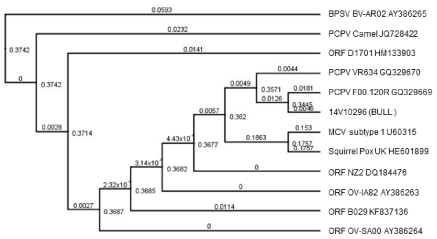
Figure 5 : Phylogenetic analysis of UDG DNA sequences from selected parapoxviruses. The tree was rooted with a BPSV BV-AR02 (AY386265), and the 14V10296 isolate UDG DNA sequence at 181bp was compared to 180 bp of selected parapoxviruses including BPSV BV-AR02, PCPV camels, PCPV VR634, PCPV F00.120R, Molluscum contagiosum virus (MCV) subtype 1, squirrel pox virus, and Orf viruses including ORF NZ2, ORF B029, ORF OVIA82, ORF D1701 and ORF OV-SA00. The bootstrap values are designated at branching nodes. The branch labels are the genetic distance (nucleotide substitutions per site).
The viral encoded IL-10 is often acquired from their host during co-evolution. To determine the 14V10296 isolate’s relationship to other PCPV viruses, the viral IL-10 PCR product amplified by primers PCP-IL10F356 and PCP-IL-10R356 was sequenced, and obtained a 329 bp sequence (Gen Bank accession No. KM277583). Comparing to other Parapoxvirus sequences, the vIL-10 of the 14V10296 isolate is also found to be closest to PCPV-F00.120R, which was isolated from reindeer in Finland between 1999 and 2000 (Table 2 and Figure. 6). The vIL-10 DNA of the 14V10296 isolate has only 73.6% to 76% homology to that of Orf virus and BPSV, respectively (Table 2). In agreement with UGD DNA sequence analysis, the vIL-10 of 14V10296 isolate is also closely related to PCPV-F00.120R (Figure. 6).
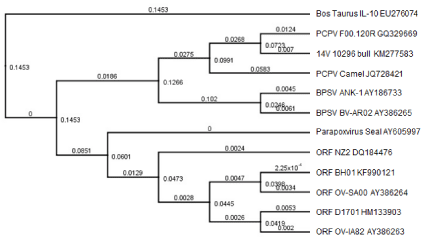
Figure 6 : Phylogenetic analysis of IL-10 DNA sequences from selected PCPV viruses. The tree was rooted with the IL-10 of cattle (Bos taurus) (EU276074), and the 14V10296 isolate IL-10 DNA sequence (329 bp) (GenBank accession No. KM277583) was compared to selected parapoxviruses including PCPVF00.120R, PCPV-Camel, BPSV Ank, BPSV BV-AR02, Seal parapoxvirus, ORF NZ2, ORF BH01, ORF OR-SA00, ORF D170 and ORF OV-IA82. The bootstrap values are designated at branching nodes. The branch labels are the genetic distance (nucleotide substitutions per site).
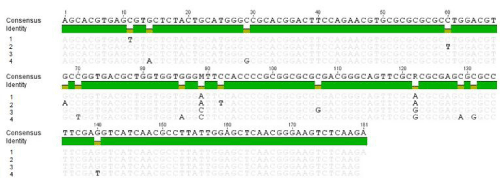
Figure 7 : DNA sequence alignments of PCR product amplified with primers specific for uracil DNA glycosylase (UDG). Lane 1: 14V10296 isolate; lane 2: PCPV F00.120R (GQ329669); lane 3: PCPV VR634 (GQ329670); lane 4: PCPV camels (JQ728422).
Discussion/Conclusion
In this report, a pseudocowpox virus (PCPV) was isolated from genital tissues of one of two bulls which failed to breed in the spring of 2014. Two bulls were examined at Rocky Mountain Large Animal Clinic in Spanish Fork, UT and were found to have poxvirus like lesions on the penis. A preputial swab was collected from one bull, but the other bull would not tolerate being swabbed and only a superficial scraping was obtained. A PCPV was isolated from the swab from one bull, but no virus was isolated from the scraping sample of the second affected bull. It is possible that the scraping sample did not contain infected material and therefore PCPV failed to be isolated. Although PCPV is a common, mild infection of the udder and teats of cows, infection of the genital tissue of cattle has not been reported. This is the first report that PCPV infects the reproductive system which may lead to new cautionary measures about the transmission and spread of PCPV infections in breeding herds.
Pseudocowpox virus infection is distributed worldwide and mostly affects milking cows [8]. The virus is usually introduced to herds through infected animals and disseminates slowly among the animals. Transmission within herds occurs by direct and indirect contact. Indirect routes include calf suckling of multiple cows, flies, milking equipment, inadequate milking/management procedures [9,10]. Most PCPV infections are associated with skin infections on the teats, udder, and foot in cattle, camels, reindeer and cats [11-14] or skin infections of human hands [15]. In addition, PCPV infection is different from BPSV infection in that BPSV infects cattle of all ages, and lesions are usually seen on the muzzles of calves [16] and less frequently on the udders and teats of cows [16]. Pseudocowpox virus can also infect humans and causes painful localized skin infections commonly known as milker’s nodules [17,18]. The protective immunity to Parapoxvirus infection in an individual is often short, despite the induction of a specific immunity and the animal can get infected fairly soon after recovering from disease, as demonstrated by Orf virus which repeatedly infects its host [19-22]. Since PCPV infection is common in milking herds, cautions should be made to prevent cross contamination to breeding herds. Subclinical infection in certain animals could lead to clinical disease in breeding herds. In addition, if a bull fails to breed, PCPV infection should now be considered as a differential diagnosis. The first documented report of pseudocowpox virus infection is from 1963 in tissue culture from bovine skin and oral lesions typical of pseudocowpox [23]. The source of the 14V10296 isolate may come from animals within the original herd with mild infection of PCPV.
PCPV uracil DNA glycosylase (UDG) gene encodes a 232 amino acid protein and has a UDG-like super family domain. The protein is important in DNA replication, recombination and repair [24]. This UDG gene seems to be conserved within parpapoxviruses. Although DNA sequences between different parpapoxviruses was found to be about 91% to 98% identical, no amino acid coding difference were found between Orf virus and PCPV or BPSV and PCPV. This suggests the UDG must be functionally important and conserved. DNA sequence analysis revealed that the UDG DNA sequence of the 14V10296 isolate is closer to PCPV-F00.120R isolate, which was isolated from Finnish reindeer (Figures 5,7) [25]. F00.120R was associated with pustular stomatitis in reindeer between 1999 and 2000. The reindeer isolate contains homologues to all known Orf virus genes. It is interesting to find this PCPV of 14V10296 isolate from Idaho is closely related to a reindeer isolate from Finland in that they both are characterized by a non-conventional distribution of lesions.
Many viruses have evolved strategies to deregulate the host immune system. They are able to acquire a cellular gene independently and evolve with their host. Many viruses were reported to encode functional orthologues of cellular IL-10, called virus IL-10s (vIL-10s). To date, vIL-10 orthologues have been reported for 12 members of the family Herpesviridae, two members of the family Alloherpesviridae, and seven members of the family Poxviridae [26]. PCPV of Poxviridae also encodes a vIL-10 gene. Based on the 329 bp sequence of the 14V10296 vIL-10 (Gen Bank accession No. KM277583), it only has about 74% homology to Orf vIL-10 and 76% homology to BPSV vIL-10, respectively (Table 2), which suggests they evolved differently and acquired the gene from different hosts. Therefore, PCR primers specific for Orf vIL-10 failed to amplify PCPV vIL-10 (Figure 3 lane 9-12). The vIL-10 DNA sequence analysis revealed that the PCPV 14V10296 bull isolate is different from the PCPV camel isolate. In agreement with UDG DNA sequence analysis, the vIL-10 of 14V10296isolate is 98.1% homologous to that of PCPV of strain F00.120R. Therefore, the 14V10296 isolate may come from a virus that is closely related to PCPV of reindeer.
Acknowledgement
This work was supported by the Department of Biomedical Sciences, College of Veterinary Medicine at Oregon State University. The EM work was supported by National Science Foundation via the Major Research Instrumentation (MRI) Program under Grant No. 1040588.
References
- Robinson AJ, Mercer AA. Parapoxvirus of red deer: evidence for its inclusion as a new member in the genus parapoxvirus. Virology. 1995; 208: 812-815.
- Abu EM, Housawi FM. Drastic cutaneous multi-focal orf infection in goats, causing severe dysfunctioning. Rev Sci Tech. 2009; 28: 1025-1029.
- Bowman KF, Barbery RT, Swango LJ, Schnurrenberger PR. Cutaneous form of bovine papular stomatitis in man. JAMA. 1981; 246: 2813-2818.
- de Sant'Ana FJ, Leal FA, Rabelo RE, Vulcani VA, Moreira CA Jr, Cargnelutti JF, et al. Coinfection by Vaccinia virus and an Orf virus-like parapoxvirus in an outbreak of vesicular disease in dairy cows in midwestern Brazil. J Vet Diagn Invest. 2013; 25: 267-272.
- Inoshima Y, Morooka A, Sentsui H. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods. 2000; 84: 201-208.
- Harkness JW, Scott AC, Hebert CN. Electron microscopy in the rapid diagnosis of ORF. Br Vet J. 1977; 133: 81-87.
- Thomas V, Flores L, Holowczak JA. Biochemical and electron microscopic studies of the replication and composition of milker's node virus. J Virol. 1980; 34: 244-255.
- Roess AA, McCollum AM, Gruszynski K, Zhao H, Davidson W, Lafon N, et al. Surveillance of parapoxvirus among ruminants in Virginia and Connecticut. Zoonoses Public Health. 2013; 60: 543-548.
- Almagro M, Maestre JR, Martínez P, Malagón I, Pérez E, Herrera I. [Milker's nodes: transmission by fomites and virological identification]. Enferm Infecc Microbiol Clin. 1991; 9: 286-288.
- Schuler G, Hönigsmann H, Wolff K. The syndrome of milker's nodules in burn injury: evidence for indirect viral transmission. J Am Acad Dermatol. 1982; 6: 334-339.
- Cargnelutti JF, Flores MM, Teixeira FR, Weiblen R, Flores EF. An outbreak of pseudocowpox in fattening calves in southern Brazil. J Vet Diagn Invest. 2012; 24: 437-441.
- Tikkanen MK, McInnes CJ, Mercer AA, Büttner M, Tuimala J, Hirvelä-Koski V, et al. Recent isolates of parapoxvirus of Finnish reindeer (Rangifer tarandus tarandus) are closely related to bovine pseudocowpox virus. J Gen Virol. 2004; 85: 1413-1418.
- Fairley RA, Mercer AA, Copland CI, Craig SM, Heslip PA. Persistent pseudocowpox virus infection of the skin of a foot in a cat. N Z Vet J. 2013; 61: 242-243.
- Abubakr MI, Abu-Elzein EM, Housawi FM, Abdelrahman AO, Fadlallah ME, Nayel MN, et al. Pseudocowpox virus: the etiological agent of contagious ecthyma (Auzdyk) in camels (Camelus dromedarius) in the Arabian peninsula. Vector Borne Zoonotic Dis. 2007; 7: 257-260.
- Falk ES. Parapoxvirus infections of reindeer and musk ox associated with unusual human infections. Br J Dermatol. 1978; 99: 647-654.
- de Sant'Ana FJ, Rabelo RE, Vulcani VA, Cargnelutti JF, Flores EF. Bovine papular stomatitis affecting dairy cows and milkers in midwestern Brazil. J Vet Diagn Invest. 2012; 24: 442-445.
- Batalla A, Alvarez-Argüelles ME, González-Martí nez MB, Curto JR. [Milker's nodule complicated with erythema multiforme]. Med Clin (Barc). 2013; 141: e5.
- Rogers M, Bale P, De Silva LM, Glasson MJ, Collins E. Giant parapox infection in a two year old child. Australas J Dermatol. 1989; 30: 87-91.
- Zhao H, Wilkins K, Damon IK, Li Y. Specific qPCR assays for the detection of orf virus, pseudocowpox virus and bovine papular stomatitis virus. J Virol Methods. 2013; 194: 229-234.
- Rohde J, Emschermann F, Knittler MR, Rziha HJ. Orf virus interferes with MHC class I surface expression by targeting vesicular transport and Golgi. BMC Vet Res. 2012; 8: 114.
- Yu YZ, Wu ZJ, Zhu ZB, Pan QZ, Cui YD. [Molecular characteristics and immune evasion strategies of ORFV: a review]. Bing Du Xue Bao. 2012; 28: 278-284.
- Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, et al. Poxviruses and immune evasion. Annu Rev Immunol. 2003; 21: 377-423.
- Moscovici c, cohen ep, sanders j, delong ss. Isolation of a viral agent from pseudocowpox disease. Science. 1963; 141: 915-916.
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011; 39: D225-229.
- Hautaniemi M, Vaccari F, Scagliarini A, Laaksonen S, Huovilainen A, McInnes CJ. Analysis of deletion within the reindeer pseudocowpoxvirus genome. Virus Res. 2011; 160: 326-332.
- Ouyang P, Rakus K, van Beurden SJ, Westphal AH, Davison AJ, Gatherer D, et al. IL-10 encoded by viruses: a remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J Gen Virol. 2014; 95: 245-262.