
Research Article
Austin Virol and Retrovirology. 2015; 2(2): 1014.
Inducing Autophagic Cell Death by Nsp5 of Porcine Reproductive and Respiratory Syndrome Virus
Liping Yang¹, Rong Wang
¹VA-MD College of Veterinary Medicine, University of Maryland, USA
²Maryland Pathogen Research Institute, University of Maryland, USA
³Laboratory Animal Center, School of Medicine, Xi’an Jiaotong University, China
*Corresponding author: Yanjin Zhang, VA-MD College of Veterinary Medicine, Maryland Pathogen Research Institute, University of Maryland, College Park, USA
Received: October 04, 2014; Accepted: November 09, 2015; Published: November 10, 2015
Abstract
Porcine Reproductive and Respiratory Syndrome (PRRS) leads to severe economic losses to the swine-producing industry. Many unclear questions remain on pathogenesis of PRRS virus (PRRSV), including the mechanism of PRRSV-induced cell death. In this study, we cloned and expressed a PRRSV non-structural protein, nsp5, and discovered that it induced cell death in cultured cells. The nsp5 protein localized in cytoplasm and majority of the protein concentrated in perinuclear region. Along with extension of incubation time, the nsp5 tended to form puncta and polarized besides nucleus. An interesting observation was that the nsp5 expression induced cell death. Cell viability assay showed that the cells with nsp5 expression had over 2-fold more cell death than cells with empty vector. Further study indicated that the nsp5 induced cell death via autophagy. Treatment with 3-MA, an autophagy inhibitor, blocked the nsp5- induced cell death. These results suggest that nsp5 might play an important role in PRRSV-induced cell death. Further examination on the mechanism is warranted.
Keywords: Porcine reproductive and respiratory syndrome virus; PRRSV; NSP5; Autophagy; Cell death
Introduction
Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) is a positive-sense single-stranded RNA virus in the family of Arteriviridae, Order Nidovirales [1]. The other members of the Arteriviridae include equine arteritis virus, lactate dehydrogenaseelevating virus, and simian hemorrhagic fever virus. PRRSV causes a contagious viral disease of pigs which is characterized by reproductive failure in sows and respiratory disease in pigs of all ages, which leads to an estimated $664 million in losses per year to the swine-producing industry in the USA alone [2]. The genome of PRRSV is a little over 15 kb in length with over ten Open Reading Frames (ORFs) [3-5]. The genome is composed of replicas genes located at the 5’-end, followed by genes encoding structural proteins. ORFs 1a and 1ab encode long polypeptides that are cleaved into individual Non-Structural Proteins (nsp), nsps 1-12, including proteases, a helicase and the RNAdependent RNA polymerase. ORFs 2-7 encode structural proteins: E, GP2, GP3, GP4, 5a, GP5, M and N.
Nsp5 is predicted to be 170 amino acids encoded by ORF1a and its predicted molecular mass is 18.9 kDa [6,7]. Nsp5 and several other nsps have been implicated in the induction of interferon-? and possibly in cell-mediated immune response [8]. Nsp5-7 was shown to cause autophagy in a similar way to nsp6 of corona viruses [9]. But the exact function of nsp5 in PRRSV replication or pathogenesis is unknown. The objective of this study to determine the expression pattern of this protein and its effect on cell viability. We discovered that nsp5 locates in cytoplasm in perinuclear region. It tends to polarize besides the nucleus along with extension of culture time. Cell viability assay showed that nsp5 causes cell death. Treatment with 3-MA rescued the nsp5-induced autophagic cell death. These results indicate that nsp5 might play an important role in PRRSV pathogenesis.
Materials and Methods
Cells, transfection, and chemicals
HEK293T, HEK293 and HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS). Transfection of the cells with plasmid DNA was performed by using FuGene HD (Promega, Madison, WI), according to the instructions of the manufacturer.
3-Methyladenine (3-MA), an inhibitor of autophagy, was used to treat cells with nsp5 expression at 5 mM.
Plasmids
The nsp5 sequence was PCR amplified and cloned to vector pCAGEN (Addgene plasmid# 11160) with HA-tag at N-terminus and VenusC1 vector as previously reported [10,11]. All primers used for plasmid construction were listed in Table 1. All constructed plasmids were subjected to DNA sequencing to confirm the inserts.
Primera
Sequence (5’ to 3’)b
Vector or use
32NSP5F1
CGGAATTCGGAGGCCTCTCCACCGTCC
pCAGEN
85NSP5R2
CCGCTCGAGTTACTCGGCAAAGTACCGCAGG
85nsp5F4
GCTCGAGGAGGCCTCTCCACCGTCC
VenusC1
85nsp5R6
AGAATTCTTACTCGGCAAAGTACCGCAGG
RR-p21-F1
ATGAAATTCACCCCCTTTCC
Real time PCR
RR-p21-R1
AGGTGAGGGGACTCCAAAGT
- F, forward; R, reverse.
- The primers for nsp5 were designed according to sequences of PRRSV VR-2332 (Gen Bank accession number U87392) and VR-2385 (Gen Bank accession number JX044140). Restriction enzyme digestion sites in primers are italicized.
Table 1: Primers used in this study.
Immunofluorescence assay (IFA)
IFA was carried out as previously reported [10], by antibody against the HA (Thermo Fisher Scientific, Waltham, MA) and DyLight 549 goat anti-mouse Immunoglobulin (Ig) G conjugate (Rockland Immunologicals, Gilbertsville, PA). The cover glass was mounted onto slide using Slow Fade Gold antifade reagent containing 4’6’-Diamidino-2-Phenylinodole (DAPI) (Invitrogen) and observed using fluorescence microscopy.
Western blot analysis
Whole proteins in cell lysate were separated by Sodium Dodecylsulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and analyzed by Western blotting as previously described [10,12]. Antibody against HA was used in the blotting. The chemiluminescence signal was recorded digitally using a ChemiDoc XRS imaging system with the Quantity One Program, Version 4.6 (Bio-Rad Laboratories, Hercules, CA).
Reverse transcription and real-time PCR (RT-qPCR)
RNA isolation, reverse transcription and real-time PCR were performed as previously described [13]. The primers used to amplify p21 and an internal control geneRPL32 (ribosomal protein L32) are listed in Table 1. Gene expression was quantified by 2-ΔΔCT method [14].
Statistical analysis
Difference of an indicator between treatment group and control was assessed by Student t-test. A two-tailed P<-value of less than 0.05 was considered significant.
Results
Nsp5 cloning and expression
Nsp5 of PRRSV VR-2385 strain was cloned into vector pCAGEN, resulting in the plasmid pCAGEN-HA-nsp5. Transfection of HeLa cells with this plasmid was conducted. IFA showed that the HAtagged nsp5 was expressed in cytoplasm of the cells (Figure 1A). Most HA-nsp5 appears in perinuclear region.
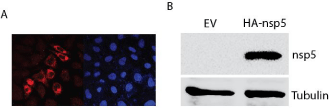
Figure 1: Detection of PRRSV nsp5 in cells transiently transfected with
pCAGEN-HA-nsp5 plasmid. A. Immunofluorescence Assay (IFA) detection of
nsp5 expression in HeLa cells. At 48 h after transfection, the cells were fixed
for IFA with antibody against HA. The nuclear is counterstained by DAPI. B.
Western Blotting (WB) detection of nsp5 expression in HEK293 cells. At 48h
after transfection, the cells were harvested for WB with antibody against HA
and tubulin.
To confirm the nsp5 expression, we transfected HEK293 cells with nsp5 plasmid and harvested the cells for Western blotting with HA antibody. Result showed that there was a specific band in an expected size of 19 kDa in HA-nsp5 lane, which was absent in the control lane with lysate of cells transfected with empty vector (Figure 1B). The results demonstrated the successful nsp5 cloning and expression.
Subcellular location of nsp5 and its temporal expression pattern
We noticed that nsp5 protein locates in the cytoplasm with some different distribution patterns. To further characterize its subcellular location and temporal expression, we cloned nsp5 into VenusC1 vector for live cell observation of the Venus-nsp5 fusion protein. HeLa cells were transfected with VenusC1-nsp5 plasmid and observed 15, 24 and 48h post transfection (hpt). The Venusnsp5 protein was found to locate in perinuclear region (Figure 2) at 15 hpt, while homologous distribution of GFP protein was observed for the cells transfected with the control vector. The distribution of nsp5 protein tends to be polarized by forming one or two big dots in perinuclear region 24 and 48 hpt. The dot became bigger 48 hpt than 24 hpt. The results indicate that nsp5 expression has a unique distribution pattern that might relate to its function.
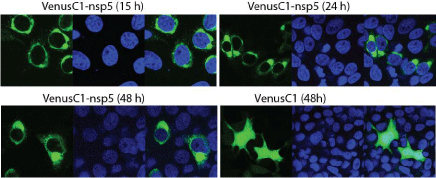
Figure 2: Nsp5 subcellular location and temporal distribution in HeLa cells
15, 24 and 48h post transfection. The cells were transfected with VenusC1-
nsp5 plasmid. At 15, 24 and 48h after transfection, the cells were fixed and
the nuclear is counterstained by DAPI before confocal microscopy. The left
image in each panel shows the Venus-nsp5 fusion protein, the middle image
shows the DAPI staining of nucleus and the right is the merged image.
Nsp5 induces cell death of HEK293 cells
In testing nsp5 expression, we noticed that cells with nsp5 expression tend to have smaller number and poorer condition than control cells (Figure 3A). So we performed cell viability assay to assess the cell death in cells transfected with nsp5 in comparison with empty vector control. Confluent monolayer of HEK293 cells was used for transfection. Result showed that nsp5 expression in HEK293 cells led to 63% reduction in cell viability compared with cells transfected with empty vector (Figure 3B). This suggests that nsp5 presence in the cells caused cell death as confluent monolayer of cells were transfected with nsp5 or empty vector.
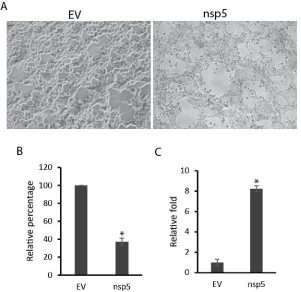
Figure 3: Nsp5 expression induces cell death. A. Phase contrast image of
HEK293 cells transfected with pCAGEN-HA-nsp5 plasmid. Empty Vector
(EV) was included as a control. B. Nsp5 induces cell death of HEK293 cells.
At 48 hours after transfection, the cells were harvested for cell viability assay.
Error bars represent standard errors of three replicates of one representative
experiment. C. Nsp5 increases p21 transcript level in HEK293 cells. At
24 hours after transfection, the cells were harvested for RNA isolation and
RT-qPCR. Error bars represent standard errors of three replicates of one
representative experiment. “*” indicate significant difference (P < 0.05) from
EV control.
To further analyze the reasons for the cell viability reduction, we conducted real-time RT-PCR to detect p21CIP1/WAF1 gene expression as it is a potent cyclin-dependent kinase inhibitor regulating cell cycle progression. Result showed that the cells transfected with nsp5 plasmid had 8.2-fold more p21CIP1/WAF1 transcript than empty vector control (Figure 3C), indicating that nsp5 induced the expression of p21CIP1/WAF1 gene. This suggests that cells with nsp5 expression tend to have growth arrest, which may precede cell death and contribute to lower cell viability than the control.
Cell death is blocked with treatment of autophagy inhibitor
As nsp5 induces cell death, we intended to figure out what mechanism it is. A previous study showed that nsp5-7 induces autophagy [9]. We reasoned that nsp5 alone might be able to induce autophagy and potentially cause cell death when excessive autophagy is beyond control. An autophagy inhibitor, 3-MA, was used to treat HEK293 cells transfected with nsp5 plasmid. A pan caspase inhibitor, ZVAD (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]- fluoromethylketone), was also included to determine if apoptosis was involved in the nsp5-induced cell death. Treatment of the cells with 3-MA led to reduced cell death in the cells with nsp5 (Figure 4). The treatment of ZVAD had no effect. This indicates that nsp5 induced cell death via autophagy and the 3-MA treatment prevented the process.
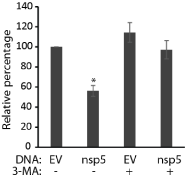
Figure 4: Treatment with 3-MA prevents nsp5-induced cell death. HEK293
cells were transfected with pCAGEN-HA-nsp5 plasmid and treated with 3-MA
at the time of transfection. Empty vector (EV) was included as a control. The
cells were harvested for cell viability assay 48 hours after transfection. Error
bars represent standard errors of two experiments. “*” indicate significant
difference (P < 0.05) from EV control.
To further confirm the nsp5 induces autophagy, we performed con-transfection of HEK293 cells with mCherryC1-LC3 and VenusC1-nsp5 plasmids. More puncta were observed in cells with nsp5 expression than empty vector (not shown). Interestingly, colocalization of nsp5 and LC3 was observed. These results suggest that nsp5 induced LC3 relocation to autophagsomes and autophagy as a result.
Discussion
This study discovered that PRRSV nsp5 protein locates in perinuclear region and induces cell death via autophagy. Among 170 nsp5 amino acid residues, 93 (55%) are hydrophobic and 34 are polar. The Protparam Program (https://web.expasy.org/protparam/) was used to analyze the nsp5 parameters. The grand average of hydrophobicity for nsp5 is 1.056, suggesting the high hydrophobic nature of this protein. Presumably, the protein is expressed in Endoplasmic Reticulum (ER) in large amount without proper folding and accumulation of mis-folded protein induces autophagy and then cell death. But this speculation needs further study to verify. On the other hand, in the presence of other viral proteins, nsp5 folding might be properly processed or its expression level may be under control.
The nsp5 was successfully cloned and expressed, which was confirmed by both IFA and Western blotting. The protein appears to have perinuclear localization and become polarized along with incubation time. The distribution pattern is consistent with its presumptive ER location. The polarized big dot may be due to accumulation of nsp5 in the ER. Indeed, a preliminary study showed that the nsp5 co-localizes with an ER marker (data not shown).
An interesting feature of the nsp5 over expression is induction of cell death. It is the main reason that we were unable to establish a stable cell line expressing nsp5. The possible reason maybe the nsp5 accumulation and misfolding in ER, which led to autophagy followed by cell death. The nsp5 induction of autophagy is consistent with previous finding that PRRSV nsp5-7 activates autophagy [9]. This property is also shared by nsp6 of corona viruses mouse hepatitis virus and Severe Acute Respiratory Syndrome virus.
The nsp5-induced cell death was prevented by 3-MA treatment to block autophagy. 3-MA inhibits autophagy by blocking autophagosome formation. The result indicates that the nsp5-induced autophagosome formation was inhibited. Although autophagy is a cellular mechanism for survival in adverse conditions, it is also known as type II programmed cell death due to accumulation of autophagosomes in the cytoplasm [15].
Conclusion
PRRSV nsp5 was shown to distribute in perinuclear region in cells and become polarized along with extension of culture time. Nsp5 induces autophagic cell death, which can be blocked by inhibition of autophagosome formation via 3-MA. Further study is warranted to elucidate the mechanism of nsp5 induction of autophagy and its contribution to PRRSV pathogenesis.
Acknowledgement
This work was partially funded by internal fund of the University of Maryland.
References
- Faaberg KS, Balasuriya UB, Brinton MA, Gorbalenya AE, Leung FC-C, Nauwynck H, et al. Family Arteriviridae. King AMQ, Elliot Lefkowitz, Michael J Adams, Eric B Carstens, editors. In: Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London. 2011.
- Wang R, Xiao Y, Opriessnig T, Ding Y, Yu Y, Nan Y, et al. Enhancing neutralizing antibody production by an interferon-inducing porcine reproductive and respiratory syndrome virus strain. Vaccine. 2013; 31: 5537-5543.
- Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993; 193: 329-339.
- Meulenberg JJ, de Meijer EJ, Moormann RJ. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993; 74: 1697-1701.
- Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol. 2011; 92: 1107-1116.
- Li Y, Tas A, Snijder EJ, Fang Y. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol. 2012; 93: 829-839.
- Li Y, Tas A, Sun Z, Snijder EJ, Fang Y. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res. 2015; 202: 48-59.
- Rascón-Castelo E, Burgara-Estrella A, Mateu E, Hernández J. Immunological features of the non-structural proteins of porcine reproductive and respiratory syndrome virus. Viruses. 2015; 7: 873-886.
- Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011; 7: 1335-1347.
- Kannan H, Fan S, Patel D, Bossis I, Zhang YJ. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol. 2009; 83: 6375-6382.
- Wang R, Nan Y, Yu Y, Zhang YJ. Porcine reproductive and respiratory syndrome virus Nsp1β inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-α1 degradation. J Virol. 2013; 87: 5219-5228.
- Zhang YJ, Wang KY, Stein DA, Patel D, Watkins R, Moulton HM, et al. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res. 2007; 73: 12-23.
- Patel D, Opriessnig T, Stein DA, Halbur PG, Meng XJ, Iversen PL, et al. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res. 2008; 77: 95-107.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402-408.
- Schwartz LM, Smith SW, Jones ME, Osborne BA. Do all programmed cell deaths occur via apoptosis? Proc Natl Acad Sci U S A. 1993; 90: 980-984.