1Institute of Biochemistry, Carleton University, Canada
2Department of Chemistry, Carleton University, Canada
3Food Science and Nutrition Program, Carleton University, Canada
*Corresponding author: Tsopmo A, Institute of Biochemistry, and Food Science and Nutrition Program, Carleton University, 1125 Colonel By Drive, K1S 5B6, Ottawa, ON, Canada
Received: August 13, 2014; Accepted: November 25, 2014; Published: November 28, 2014
Citation: Gao Q, Smith JC and Tsopmo A. Optimized Protamex Digested Oat Bran Proteins: Antioxidant Properties and Identification of New Peptides. Austin J Nutri Food Sci. 2014;2(10): 1053. ISSN: 2381-8980.
The aim of this study was to determine the radical scavenging properties and spectroscopic characteristics of hydrolysates produced from oat bran proteins digested for 1-4h and 1:10 to 1:40 enzyme (Protamex)/substrate (E/S) ratios. Four complementary bioassays were used to determine the activity of the twelve hydrolyzed proteins. The oxygen radical absorbance capacity assay showed that digestion for 3h produced hydrolysates with higher peroxyl radical scavenging activities (521-695 μM TE/g) while digestion for 2h with either 1:20 or 1:30 E/S ratio resulted in greater inhibition of superoxide radical anions 57.4±5.1% and 57.7±1.6%, respectively. The hydroxyl radical scavenging activity of all hydrolysates decreased overtime while there was little time dependent change in ferrous ion chelating activity.Overall, hydrolysis with 1:30 E/S for 2 or 3h produced hydrolysates with greater antioxidant capacity due to their relative higher content of hydrophobic (Leu+Ile+Val) and aromatic (Tyr+Phe) amino acids as indicated by 1H-NMR data. The sequence of nine novel peptides was determined by LC-MS/MS in these two most active samples.
Keywords: Antioxidant; Oat peptides; 1H-NMR; LC-MS/MS; Nutrition; Proteolysis; Protamex
The functionality and the biological activity of food protein isolates and their hydrolysates are currently the focus many investigations mainly because they contain peptides that may be used in the prevention of chronic diseases or for general well-being [1-3]. It has been shown that short-chain peptides from hydrolyzed proteins have higher nutritive values and may also be utilized more efficiently by the body than mixtures of free amino acids [4]. Protein hydrolysates are used not only in energy drinks, weight-control and sports nutrition products [1], but also in nutritional products for elderly and immuno-compromised patients [5].
Biological activities such as antioxidant, antihypertensive, hypolipidemic, hypocholesterolemic and antitumor have been attributed to peptides from enzymatically digested food proteins [3]. Antioxidants added to foods increase their stability while dietary consumption can supplement the endogenous defense systems therefore, helping maintain the redox balance (i.e. prevent oxidative stress) [6]. Anti-oxidative properties of peptides from foods like milk, soy, fish, quinoa, linseeds exist in the literature [2,3].
Oats contain 15-20% proteins that possess better nutritional value compared to other cereals because of their higher content of the essential amino acid lysine [7]. The determination of biological activities of digested oat proteins is therefore of importance. It has recently been shown that oat bran proteins digested with alcalase and trypsin released new peptides and possessed antioxidant properties [8], however the use other proteases haven't been investigated. Protamex is an endopeptidase with a broad specificity to hydrophobic amino acids that has been used to produce bioactive protein hydrolysates from foods like rice [9]. There is however no literature information for its utilization on oat proteins. The objectives of this study were then to evaluate the effect of digestion time and Protamex-protein isolate ratio on the release of peptides (i.e. amino acid composition and sequencing) from oat bran; to determine the antioxidant activities of the protein hydrolysates; and to characterize the digests by proton nuclear magnetic resonance and mass spectrometry.
Medium oat bran flour (item number 112-001) was supplied by Can-Oat Milling Inc. (Portage La Prairie, MB, Canada). Viscozyme L 100 Fungal Beta Glucanase (FBG)/g, potassium dichromate, pyrogallol, Tris-HCl, Ethylene Diamine Tetra-Acetic (EDTA), sodium phosphate monobasic dehydrate, 1,10-phenanthraline, glutathione, mono- and dibasic potassium phosphates were obtained from Sigma-Aldrich (Oakville, ON, Canada). Methanol, hexane, hydrogen peroxide, and ferrous sulfate heptahydrate were purchased from Fisher Scientific Co. (Nepean, ON, Canada). 2,2'-azobis(2- methylpropionamidine) dihydrochloride (AAPH) was obtained from Wako Chemical (Richmond, VA, USA). Spectrophometric measurements were performed on 96-wells BioTek® Epoch™ UV-Vis or BioTek® FLx800 fluorescence readers both controlled by Gen5™ data analysis software while incubations were done on a MaxQ™ 5000 incubator shaker model (Fisher Scientific, Nepean, ON).
The extraction of proteins was based on a reported procedure that used Viscozyme, a multi-enzyme complex to decrease the viscosity of the slurry (breakdown of carbohydrates) and improve protein yield because of better precipitation during centrifugation [8]. Briefly, Defatted brans (3x80 g grams) were mixed with water 1:10 (w/v) followed by adjustment to pH 4.5. Viscozyme (3 FBG/g) was then added followed by incubation at 45°C, 150 rpm for 2h. At the end of the incubation, the pH was adjusted to 9.5 with 2 M NaOH solution and further incubated for 30 min. Alkaline solutions were cooled and centrifuged at 2500g, 20 min and 4°C. Protein isolates were precipitated after centrifugation of supernatants (pH 4.0) at 10,000g for 40 min at 4°C. The isolates were dissolved in water adjusted to pH 7.0 and freeze dried.
Oat bran protein isolate (800 mg each) was transferred to sixteen different tubes and suspended in water 1:12 ratio. The pH was adjusted to 6.5 with 1 M NaOH and 1M HCl followed by addition of Protamex at 1:10, 1:20, 1:30 and 1:40 (w/w) Enzyme-Substrate (E/S) ratios. The hydrolysis was performed at 50°C and 150 rpm. Four samples were removed at 1, 2, 3 and 4 h, respectively. At the end of each incubation time, the enzyme was inactivated by heating at 90°C for 10 min and the tubes were allowed to cool at 4 °C. Supernatants were collected after centrifugation at 5000g for 25 min to remove enzymes and non-digested proteins, freeze dried and stored at - 20°C until further analyses. The Amino Acid (AA) composition of the freeze dried digested proteins was performed by Advanced Protein Technology Centre at the Hospital for Sick Children, Ontario, Canada. Picomoles of individual amino acids detected were converted to percentage relative to total amino acids content.
This assay was performed according to literature [10]. Potassium phosphate buffer (75 mmol/L, pH 7.4) was used to prepare all reagents, standards, samples, and the control. Five trolox concentrations between 6.25 and 100 μmol/L were used for calibration curve. Fluorescein (0.080 μmol/L) and AAPH (0.12 mol/L) were prepared immediately before use. Two concentrations (50 and 100 μg/mL)of protamex digested oat bran proteins and Glutathione (GSH, control) were used in triplicate. Data were collected every min for a total of 50 min and processed as described by Cao and Prior [11]. ORAC values were expressed as Trolox Equivalents (TE) using the standard curve.
The superoxide scavenging activity of the digested oat proteins and GSH was measured as previous reported [12]. Hydrolysates and GSH were dissolved in 50 mmol/L Tris-HCl buffer containing 1 mM EDTA, pH 8.3 to a final concentration of 1mg/mL. To run the assay 80 μL of each digested oat bran protein or GSH control was mixed with 80 μL of buffer in a 96-well clear round bottom microplate in darkness. Then, 70 μL of 1.5 mMpyrogallol dissolved in 10 mmol/L HCl was added. The reaction rate (ΔA/min) was measured immediately at 420 nm for 4 min at room temperature on the BioTeK microplate reader. The buffer was used as a negative control. The superoxide scavenging activity was calculated using the following equation
Scavenging activity (%) = ((ΔA/min)c - (ΔA/min)s)/(ΔA/min)c)x100
Digested proteins were prepared in 0.75 mmol/L potassium phosphate buffer at pH 7.4. Then 50 μL of each solution were added to 96-well clear microplate in triplicate followed by the addition of 50 μL of 3 mmol/L 1,10-phenanthroline in phosphate buffer and 50 μL of 3 mmol/L FeSO4.7H2O in water. To initiate the reaction, 50 μL of 0.03% aqueous H2O2 was added, and the plate was sealed and incubated at 37 °C for 1 h with shaking (120 rpm). The HO· scavenging activity in percentage was calculated using the following formula [(As - Ac)/ (A0 - Ac)] x 100. As: absorbance of the sample; Ac: absorbance of the control solution containing 1,10-phenanthroline, FeSO4 and H2O2; A0: absorbance of the blank solution containing 1,10-phenanthroline and FeSO4.
The metal chelating assay was based on a previous method [13] with slight modifications. An aliquot (0.5 mL) of digested oat proteins or GSH (0.5 or 1.0 mg/mL) was combined with 25 μL of FeCl2 solution (2 mmol/L) and 0.9 mL of water. FerroZine™ (3-(2-pyridyl)- 5,6-diphenyl-1,2,4-triazine-4',4''-disulfonic acid sodium salt) solution (50 μL, 5 mmol/L) was added and mixed vigorously. The mixture was left to stand at room temperature for 10 min. The absorbance was measured at 562 nm. In the control, the sample was replaced with water. The chelating effect was calculated by the following equation:
Chelating effect (%) = ((Ac- As)/Ac) x 100
1H NMR spectra were obtained using a Bruker Avance 300 MHz nuclear magnetic spectrometer (Billerica, MA, USA). 10 mg of oat bran proteins digested with Protamex for 2 h and 1:10 to 1:40 E/S ratios were dissolved in 0.6 mL Deuterated water (D2O) and then transferred to NMR tubes. Spectra were recorded at room temperature with the number of scan set to 64 and were processed using TopSin version 1.3. The residual HOD signal was used for calibration.
Protein hydrolysates at 0.5 mg/mL in 1% acetic acid were injected onto a 7 cm x 200 μm inner diameter trap column, fritted, with 5 cm of 3 μm Magic C18AQ reversed phase packing material (Michrom Bioresources, Auburn, CA). The trap column was connected in series to a 6 cm x 75 μm Picofrit analytical column (New Objective, Woburn, MA). Peptides were separated using a gradient of 2% B at 0 min, 5% B at 3 min, 10% B at 8 min, 29% B at 70 min, 80% B at 75 min. A and B are 0.1% formic acid/water and 0.1% formic acid/acetonitrile, respectively. Peptides were eluted from the trap and analytical columns at a flow rate of 250 nL/min, ionized by nano Electro Spray Ionization (ESI) and analyzed using a QSTAR XL QqTOF mass spectrometer (AB Sciex, Concord, ON) over a mass range of 300 to 2000 m/z. Only ions with charge state 2+ and 3+in the MS scan were automatically selected for collision-induced fragmentation (MS/MS). Data were analyzed using Mascot (Matrix Science Ltd, Boston, MA). Mass tolerances were set to ± 100 ppm and ± 0.6 Da for the peptide and fragment ion spectra, respectively; one missed cleavage was considered; no modification was selected; enzyme was to none. Peptides identified by Mascot were manually examined and verified for accuracy.
Data are expressed as mean ± standard deviation from replicates (n = 3). ANOVA was performed and when significant (P <0.05), means were separated using Fisher's protected Least Significant Difference (LSD) test (alpha level = 0.05).
Four assays were used to evaluate the antioxidant activity of oat bran proteins digested with four concentrations of the endoprotease Protamex coupled with four digestion times (1-4h) to produce sixteen hydrolysates. The assays chosen are specific to common oxidant species like transition metal ions; peroxyl, superoxide anion and hydroxyl radicals that are responsible for damage of food and biological molecules. The hydrolysates displayed various activities that were compared to the activity of GSH, an important anti-oxidative peptide.
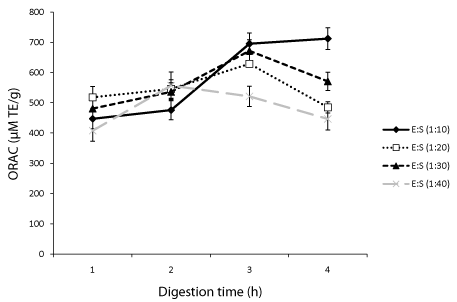
ORAC is widely employed to assess the free radical scavenging activity of protein hydrolysates and other food molecules [3]. In this study, it was used to evaluate how Protamex concentration and digestion time affect the scavenging activity of different oat bran protein hydrolysates. Results showed that at 1:10 (E/S) ratio, the scavenging activity increased with increasing digestion time indicating that smaller peptides had greater contributions than larger ones. At this E/S ratio (1:10), the hydrolysates obtained after 1 h had an ORAC value of 447±33 μM Trolox Equivalents (TE)/g of sample compared to 476±32, 695±36 and 712±36 μM TE/g for 2, 3 and 4 h, respectively (Figure 1). Digestion performed with 1:20 and 1:30 E/S ratios showed that ORAC increased from 1 to 3 h but decreased after 4 h digestion (Figure 1). At each digestion time, the peroxyl radical scavenging activity of oat bran proteins treated with 1:40 (E/S) ratio was generally lower compared to the activity at other E/S ratios.
A previous investigation showed that the E/S ratio and the duration of digestion had positive effect on ORAC values of hydrolyzed milk alpha-lactalbumin, caseino-macropeptide and whey protein concentrate [14]. Data from this study have also demonstrated the positive effect of the E/S ratio and hydrolysis time on peroxyl radical scavenging activity. Specifically with the same E/S ratio the ORAC value increased up to 3 h digestion except for 1:40 E/S ratio where the activity started decreasing after 2 h. For proteins digested for 3 and 4 h, we found that the activity of the hydrolysates increased with increasing concentration of Protamex. Glutathione, an antioxidant peptide used as standard was about 2 fold more active than the best performing hydrolysate.
The increased in peroxyl radical scavenging activity of hydrolyzed proteins has been linked to the release of specific peptides that can donate protons to radical species therefore, making them un reactive [3]. Many reports have shown that amino acids like tryptophan and tyrosine both containing aromatic ring moieties can form relatively stable radical intermediates that in turn can break radical chain reactions [15]. In peptides that contain the amino acid methionine, its sulfur moiety can be preferentially oxidized and therefore help prevent or reduce oxidative damage to foods components [16]. The length of peptides and the position of amino acids on the sequence are also important for the activity [17]. The Amino Acid (AA) composition of the oat bran proteins treated with 1:20 E/S and hydrolyzed for 1-4h as well as for proteins hydrolyzed for 2h with the four E/S ratios was obtained (Table 1) to see if there was any correlation with antioxidant activities. The analysis showed that there were small variations in composition of aromatic AAs, positively charged AAs, and negatively charged AAs. For proteins treated with 1:20 E/S ratio and digested for 1-4h, the highest ORAC value associated with 3 h digestion may be explained by a slightly higher percentage of aromatic and negatively charged AAs that have the potential to form stabilize radical intermediates [15]. It may also be explained by the high combined percentage of proline, histidine and tyrosine as they are important residues in radical scavenging properties of peptides. The ORAC values (408-712 μM TE/g) of hydrolysates obtained here are higher that values previously reported for oat bran proteins digested with alcalase [8] but comparable to those obtained for trypsin, pepsin and flavourzyme rapeseed protein hydrolysates [19]. They are however lower than values of some milk hydrolysates (63-1489 μM TE/g) [20].
Superoxide anion radical (O2·−) created by addition one electron to molecular oxygen is a harmful species to cellular components when produced in excess. In the presence of antioxidants, O2·− radicals are scavenged leading to the decrease in rate of pyrogallol (assay probe) autoxidation. The highest activity (Figure 2) was associated with proteins digested for 2h and 1:20 E/S (57.4±5.1%) or 1:30 E/S ratio (57.7±1.6%). The scavenging activity of samples digested with 1:40 E/S ratio decreased after 1h and remained constant between 2-4h probably because the enzyme was entirely consumed after 1h.
Investigation of the effects of hydrolysis time (0.5 - 4 h) and protease type on the antioxidant activity of the barley storage protein found a slight decrease with no clear relationship between O2·− scavenging activities and time for hydrolysates obtained with flavourzyme, alcalase or pepsin [21]. It was then concluded that the lack of relationship between O2·− scavenging activity and hydrolysis time was an indication that the specific cleavage site and peptide sequences were more important than the time. In this study, we found that O2·− activity only decrease over time with oat bran proteins hydrolyzed with 1:40 E/S ratio. For protein isolates treated with 1:10, 1:20 and 1:30 E/S ratios the highest O2·− activities were obtained after 2 h digestion (50.2 - 57.7% inhibition). A decrease to 25.9-33.6% and 23.5-27.4% inhibition range was observed after 3 and 4 h digestion, respectively. The AA composition of oat bran proteins digested for 2 h could not be used to explain differences in proteins treated with differences E/S ratios suggesting as found in other studies [17] that other factors such the length and sequence of peptides had greater contribution. The O2· activity for 2 h and 1:20 or 1:30 E/S ratios oat protein hydrolysates (57.4, 57.7%) were higher than those obtained for rapeseed proteins hydrolysates and their membrane-separated fractions but comparable to soy protein hydrolysates values [22].
The Hydroxyl radical (HO·) is the most reactive oxygen species formed in biological systems and as such reacts rapidly with sugars, amino acids, phospholipids, DNA bases and organic acids found in living cells [23,24]. The removal of HO· radicals can therefore reduce cell damages and possibly protect humans against many chronic diseases [25]. The 1,10-phenanthroline/Fe2+/H2O2 system was used in this study to determine the HO· scavenging capacity of oat bran protein hydrolysates and data are shown in Figure 3. Oat bran proteins digested for 1h with 1:10, 1:20, and 1:30 E/S ratios had significantly higher HO· scavenging activities (10.4-11.3% inhibition) compared to activities at any other digestion time and E/S ratio. It was followed by sample digested for 2h with 1:30 E/S ratio. At each E/S ratio the HO· scavenging activity decreased from 1 to 4h. Linear correlation coefficients (R2) were 0.91, 0.76, 0.92, and 0.98 for proteins digested with 1:10, 1:20, 1:30, and 1:40 E/S ratio, respectively. The HO· scavenging activities obtained here are lower than those of alcalase hydrolysates from oat bran (9.7-28.6%) [8] and rice endosperm proteins [26].
Transition metal ions such as iron can catalyze certain chemical reactions and generate radicals that trigger oxidation chain reactions in biological and food systems [27]. The chelation of Ferrous ions (Fe2+) which may be accomplished through electrostatic interaction with charged AA residues or through peptide transition metal ion entrapment will decrease the amount of free iron available to participate in the Fenton reaction and ultimately decrease the formation of HO· radicals [26]. As shown in Figure 4, at each digestion time the chelating ability of digested oat bran proteins increased with decreasing Protamex concentration. At each E/S ratio, proteins digested for 1h had the highest (P < 0.05) chelating activity. There was a moderate linear correlation (R2 = 0.66) between the activity and the percentage of negatively charged AA for proteins digested for 2h. It appears that either 1:30 or 1:40 ratio E/S ratio can be used to obtain chelating peptides from Protamex digested oat bran proteins with minimal effect of the duration of hydrolysis.
Bamdad et al [21]. Found that the chelating ability of alcalase and pepsin digested barley protein increased significantly in the first hour of hydrolysis and then decreased sharply making them suggested that the relatively higher molecular weight peptides in the 1 h digested sample was responsible for ferrous ion chelating activity of hydrolysates. In this study lower activity associated with higher Protamex concentration (1:10 E/S) may then be due to the fact that their hydrolysates contain peptides of lower molecular weight.
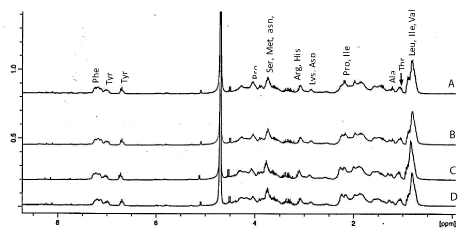
The NMR spectroscopy was further used to characterize hydrolysates from proteins digested for 2 h and different E/S ratios (Figure 5). This technique has been applied to the analysis of acid and enzyme hydrolyzed food proteins [28,29]. Signals were assigned based on the literature as it has been demonstrated that distinct amino acids have distinct 1H-NMR spectral peaks [29,30]. In the aromatic region (Figure 5), the peak for protons at about 7.3 ppm was assigned to Phe residue while those at 7.1 and 6.75 ppm were attributed to Tyr. Protons of alpha-carbons appeared between 4.01-4.38 while methyl groups for Val, Leu and Ile were centred around 0.98 ppm. It has been shown that in the proton NMR spectrum, sharp signals arise from single amino acids or short peptides while broad and distorted signals are a result a larger peptide or proteins which possessed many disordered structures and intra- or inter-molecular hydrogen bonding [30]. In this study, the better resolution of signals associated with Tyr, Val, Leu and Ile in 1:10 and 1:20 E/S ratio hydrolysates compared to corresponding signals in samples treated with 1:30 and1:40 E/S ratios may then be due to the presence of more small peptides containing these amino acids in the presence of higher Protamex concentration. The intensity of peaks normalized based of residual proton of HOD revealed that the hydrolysate from proteins treated with 1:20 E/S possessed the highest amount of combined hydrophobic (Leu + Ile + Val) and aromatic (Tyr + Phe) amino acids content. This observation can further explained the fact that this particular hydrolysate had good activity in most the assays.
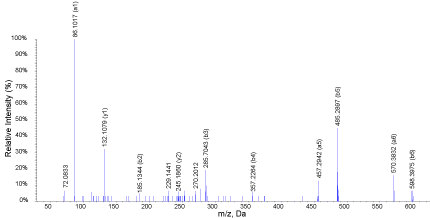
Two of the most active hydrolysates in the majority of assays were subjected to liquid chromatography tandem mass spectrometry. MS/MS data of double and tripled charged ions were then converted to peak lists that were subjected to identification using Mascot online server. MS scan of the peak eluting between 14.51-14.98 min is shown in Figure 6 in oat bran digested for 2 h and 1:30 (E/S) ratio..There are several peaks but only those with m/z 365.24 and 569.36 were double charged and automatically selected for MS/MS analysis. When peptides are double the difference between two consecutive isotopic peaks is 0.5 (e.g. m/z 365.24, 365.74, 366.22). The product ions spectrum of m/z 365.24 is showed in Figure 7. Important peaks were b-ions that extend from the amino terminus and y-ions that extend from the carboxy terminus. In MS/MS spectra they are the most common fragments. Peaks known as a-ions and derived the loss of carbonyl groups (i.e. 28 Da) from b-ions were also present derived from the loss in the spectrum. Some of the useful peaks for identification were observed at m/z598.36 (b6, [M-Ile]+), 485.27 (b5, [b6-Ala]+), 357.21 (b4, [b5-Ile]+), 286.18 (b3, [b4-Ile]+), 185.13 (b2, [b3-Ile]+). Other useful signals were a- or y-ions at m/z 444.28 (y4), 373.24 (y5),132.10 (y7), 457.29 (a5), and 570.38 (a6). The sequence elucidated as IATAQII (Ile-Ala-Thr-Ala- Ile-Ile) was present in both 2 and 3 h samples digested with 1:30 E/S ratio. Three other peptides identified in both samples were AIFSTT (m/z 320.18, 2+), KQEALAALK, (m/z 486.27, 2+), and MPIIERVIQP (m/z 598.33, 2+). Peptides QVNFTPNWGT (m/z 582.32, 2+) and LSGAQYQQHGRALK (m/z 519.62, 3+) were identified only in the 2 h digested proteins while VIGGDEDNINE (m/z 587.79, 2+), NSATTSC (m/z 342.16, 2+) and KSAGTTVMNP (m/z 503.25, 2+) were only found in those digested for 3 h. These peptides are characterized for the first time.
Data from this study have demonstrated that digestion for 2 or 3 h with 1:30 E/S ratio produced hydrolysates with stronger radical scavenging properties. The two samples also had higher content of hydrophobic and aromatic amino acids as indicated by 1H-NMR data that may account for the their better performance. Nine new peptides were found in in the two most active hydrolysates. Future studies will evaluate the bioactivity of identified peptides in food systems and cell culture models.
This work was supported by a grant from National Science and Engineering Research Council of Canada.
Amino acid composition of oat bran protein hydrolysates digested Protamex 1:20 (w/w) E/S ratio for 1-4 h and for hydrolysates digested for 2 h with 1:10 to 1:40 E/S ratio.
Amino acid |
1:20 E/S |
2 h |
||||||
1 h |
2 h |
3 h |
4 h |
1:10 E/S |
1:20 E/S |
1:30 E/S |
1:40 E/S |
|
Asx |
6.52 |
6.24 |
6.16 |
6.20 |
6.73 |
6.24 |
5.97 |
5.92 |
Glx |
29.80 |
30.41 |
30.60 |
30.13 |
29.34 |
30.41 |
31.11 |
31.88 |
Ser |
6.01 |
6.17 |
6.21 |
6.06 |
5.86 |
6.17 |
6.26 |
6.62 |
Gly |
4.77 |
4.97 |
4.99 |
4.91 |
4.87 |
4.97 |
5.01 |
5.16 |
His |
2.01 |
2.04 |
2.03 |
2.06 |
2.04 |
2.04 |
1.95 |
1.84 |
Arg |
6.98 |
6.58 |
6.46 |
6.59 |
6.73 |
6.58 |
6.44 |
6.19 |
Thr |
4.04 |
4.18 |
4.21 |
4.18 |
3.98 |
4.18 |
4.22 |
4.49 |
Ala |
4.10 |
4.03 |
4.02 |
4.11 |
4.23 |
4.03 |
3.99 |
3.80 |
Pro |
5.60 |
5.69 |
5.86 |
5.66 |
5.75 |
5.69 |
5.58 |
5.49 |
Tyr |
5.26 |
5.28 |
5.37 |
5.23 |
5.26 |
5.28 |
5.30 |
5.51 |
Val |
4.96 |
4.76 |
4.70 |
4.94 |
5.08 |
4.76 |
4.75 |
4.57 |
Met |
1.09 |
1.03 |
1.01 |
1.05 |
1.21 |
1.03 |
0.99 |
0.88 |
cys |
0.31 |
0.30 |
0.31 |
0.34 |
0.31 |
0.30 |
0.26 |
0.23 |
l l e |
3.45 |
3.32 |
3.28 |
3.44 |
3.59 |
3.32 |
3.30 |
3.17 |
Leu |
6.51 |
6.38 |
6.26 |
6.52 |
6.54 |
6.38 |
6.27 |
5.85 |
Phe |
6.07 |
6.05 |
5.98 |
6.02 |
5.87 |
6.05 |
6.05 |
5.89 |
Lys |
2.52 |
2.57 |
2.56 |
2.56 |
2.61 |
2.57 |
2.55 |
2.52 |
HAA |
32.08 |
31.56 |
31.41 |
32.09 |
32.59 |
31.56 |
31.19 |
29.88 |
AAA |
11.33 |
11.33 |
11.35 |
11.25 |
11.13 |
11.33 |
11.35 |
11.40 |
PCAA |
11.51 |
11.19 |
11.06 |
11.21 |
11.38 |
11.19 |
10.94 |
10.54 |
NCAA |
46.37 |
47.00 |
47.17 |
46.56 |
45.91 |
47.00 |
47.56 |
48.91 |
ASX: Aspartic Acid And Asparagine; GLx: Glutamic Acid and Glutamine; HAA: Hydrophobic Amino Acids (alanine, valine, isoleucine, leucine, proline, methionine and cysteine); AAA: Aromatic Amino Acids (phenylalanine and tyrosine); PCAA: Positively Charged Amino Acids (arginine, histidine, lysine); NCAA: Negatively Charged Amino Acids (Aspartic/asparagine, Glutamic/glutamine, threonine, serine).
Effect of digestion time and Enzyme Substrate ratio (E/S) on the peroxyl radical scavenging activity of oat bran proteins treated with Protamex from the Oxygen Radical Absorbance Assay (ORAC).
Superoxide anion radical (O2·-) scavenging activity of oat bran proteins digested with Protamex for 1-4 h and 1:10 to 1:40 Enzyme/Substrate (E/S) ratios.

Influence of digestion time and Enzyme Substrate ratio (E/S) on the Hydroxyl radical (HO·) scavenging activity of oat bran Protamex digested proteins.

Ferrous ions (Fe2+) chelating activities of oat bran proteins digested with Protamex for 1-4 h and 1:10 to 1:40 Enzyme/Substrate (E/S) ratios.

1H-NMR spectra of oat bran protein digested for 2 h with Protamex and 1:10 (A), 1:20 (B), 1:30 (C) and 1:40 (w/w) enzyme/substrate ratios.
TOF MS scan of molecules eluting between 14.5-15.0 min inoat bran digested for 2 h and 1/30 (Protamex/hydrolysate) ratio. Enhanced resolution showed that the signal atm/z365.24 was double charged (mass difference between isotopic peaks of 0.5 Da).
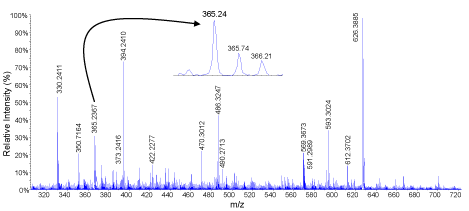
Product ions spectrum of the signal at m/z 365.24. The sequence was determined as Ile-Ala-Thr-Ala-Ile-Ileand was found in both 2 and 3 h samples digested with 1:30 E/S ratio.