
Mini Review
Austin Alzheimers J Parkinsons Dis. 2014;1(3): 4.
Tau mRNA: A Brief History of its Localization
Gonzalo Emiliano Aranda-Abreu1*, María Elena Hernández Aguilar1, Fausto Rojas Durán1 and Jorge Manzo Denes2
1Cuerpo Académico de Neuroquímica, Centro de Investigaciones Cerebrales/Universidad Veracruzana
2Cuerpo Académico de Neurociencias, Centro de Investigaciones Cerebrales/Universidad Veracruzana
*Corresponding author: Gonzalo Emiliano Aranda-Abreu, Cuerpo Académico de Neuroquímica, Centro de Investigaciones Cerebrales/Universidad Veracruzana, C.P. 91190, Xalapa Veracruz, Mexico.
Received: September 15, 2014; Accepted: October 14, 2014; Published: October 15, 2014
Abstract
We describe how tau mRNA is localized to the axon hillock and along the neuronal axon. We also reveal how proteins binding the 3’UTR region were discovered and how it was determined the way these proteins interact with tau mRNA, forming a ribonucleoprotein complex, as well as how this complex is stabilized, anchored, localized and translated in situ. Tau’s was the first mRNA molecule for which the mechanism of localization to the axon and its major function in maintenance of neuronal polarity were described.
Keywords: mRNA; Tau; Ribonucleoproteins; Subcellular localization
Introduction
An mRNA consists of three main regions: a 5’-untranslated region (5’UTR), a coding region and a 3’-Untranslated Region (3’UTR). The degradation of an mRNA highly depends on the size and sequence of the 3’UTR. The degradation process of an mRNA is tightly regulated and does not represent a random process [1]. When an mRNA leaves the nucleus, it is translated and degrades according to the particular degradation signals present in the 3’UTR. Sequences such as UUAUUUAUU [2] are sufficient for an mRNA to quickly degrade in the cytoplasm. It has also been determined that AU-regions in c/v-fos are sufficient to destabilize the mRNA molecule [3]. Expression of a number of proto-oncogenes, cytokines and lymphokines is regulated during degradation by a junction protein called HuR [4].
In neurons, an interesting event takes place, going from stabilization to the localization of several mRNAs [5,6]. Neuronal localization of messenger ribonucleic acids (mRNAs) was first studied through in situ hybridization experiments for vasopressine [7], proopiomelanocortin [8], preproenkephalin [9] and oxytocin [10], among others.
Axonal localization of tau mRNA
The study of subcellular mRNA localization began with Hirokawa’s studies on tau mRNA during brain development in the rat, in 1991 [11].
Neurons typically consist of a cell body or soma, dendrites and an axon, and thus there was a need to identify those molecules involved in cell polarity. Previous observations had shown that at least two proteins participated in neuronal polarity: tau and MAP2 [12]. Immunohistochemical analyses revealed protein tau localized to axons and MAP2 was confined to dendrites [12]. Ginzburg’s studies using in situ hybridization demonstrated the segregation of tau mRNA to the axon during neuronal development [13] and its association to microtubules [14]. Protein tau has the main function of stabilizing microtubules, thus contributing to the maintenance of neuronal polarity in the axonal region [15].
The essential question then was: how is the mRNA localized to the axon? There was the possibility that a signal sequence directed the cellular transport while preventing its degradation. Tau 3´UTR has a length of 3,865 bases. The mRNA is quite stable, with a half-life of 20h [16]. Using UV-crosslinking, it was demonstrated that two proteins of unknown nature, with molecular weights of 38 and 43 kDa, bind to a 91 nucleotide sequence in the 3’UTR of tau mRNA [17]. It was hypothesized that these proteins were necessary for the stabilization of tau mRNA during transition from neuroblasts to post-mitotic neurons [18]. Nevertheless, it had to be proved that these proteins bind naturally to this nucleotide sequence.
the axon? There was the possibility that a signal sequence directed the cellular transport while preventing its degradation. Tau 3´UTR has a length of 3,865 bases. The mRNA is quite stable, with a half-life of 20h [16]. Using UV-crosslinking, it was demonstrated that two proteins of unknown nature, with molecular weights of 38 and 43 kDa, bind to a 91 nucleotide sequence in the 3’UTR of tau mRNA [17]. It was hypothesized that these proteins were necessary for the stabilization of tau mRNA during transition from neuroblasts to post-mitotic neurons [18]. Nevertheless, it had to be proved that these proteins bind naturally to this nucleotide sequence.
Previously, it had been demonstrated that MAP2 is localized to dendrites and that its mRNA possesses a signal sequence of 640 nucleotides in the 3’UTR [19]. Similarly, the Hu proteins, which are highly conserved in vertebrates [20] and have been associated with neurological disorders due to their important roles in development and neuronal maintenance [21], bind in vitro to an AU-rich sequence and regulate mRNA degradation [22]. Furthermore, a member of the Hu family, HuD, contains three copies of RNA recognition motifs (RRM) [23,24]. Our group investigated whether HuD had the ability to bind to the 3’UTR in tau mRNA, as this protein shows a molecular weight of 38-43 kDa which is consistent with the previous findings from the UV-crosslinking experiments. Using two different cell lines: the rat pheochromocytoma PC12 cells [25] and the mouse embryonic carcinoma P19 cells [26], we proved HuD interacts with tau mRNA and determined its localization (Figure 1).
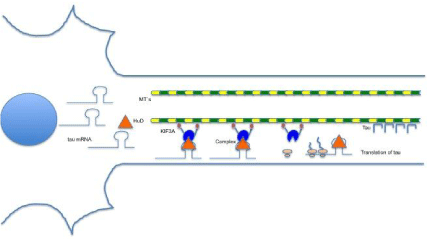
Figure 1: In the scheme shown as tau mRNA interacts with various molecules for transport to the axon. The HuD protein binds to a specific region of uracils in the 3’-UTR for this complex is subsequently anchored to the kinesin KIF3A and transported on the microtubules. Arriving at the right place, the complex disassembles the messenger of tau for translation.
In PC12 cells, we demonstrated HuD was localized to processes emanating when cells were treated with nerve growth factor (NGF) [27] and, through UV-crosslinking, its binding to an uracil-rich region in tau 3’UTR. In P19 cells differentiated with retinoic acid, we observed a phenotype characteristic of a neuronal cell with the capacity for neurotransmitter release [28]. Upon transfection of a tau uracil-rich sequence coupled to green fluorescent protein (GFP), fluorescence was observed along the axon.
After, we developed a method to immunoprecipitate (Figure 2) the complex formed by HuD and tau 3’UTR using anti-HuD antibodies, purified by Chung et al. [29], in PC12 cells in culture. We also amplified those regions near the binding site by RT-PCR.Mutations to this site prevented immunoprecipitation of the complex and therefore the amplification of regions near the binding site. With these experiments, we demonstrated HuD binds naturally to this uracil-rich site concluding that HuD proteins were able to stabilize tau mRNA and keep it in the cytoplasm for about 20h.
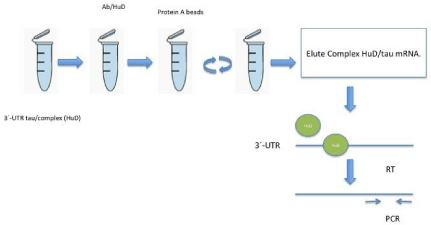
Figure 2: In this scheme briefly shows how complex formation HuD and tau messenger by immunoprecipitation and RT-PCR was determined.
Moreover, in P19 cells we showed the co-localization of tau mRNA and tau protein in the axon [30]. A group of proteins called kinesins had been identified in the squid’s giant axon [31] and Hirokawa’s studies on the motor patterns of kinesins along the microtubules [32,33] lead us to hypothesize that the HuD-tau mRNA complex was anchored to a kinesin. We proved this hypothesis using antibodies, showing that kinesin KIF3A anchors HuD and transports the HuD-tau mRNA complex to the axon [34]. When KIF3A expression was inhibited, the complex did not reach the axon. We had then completed the puzzle on how tau mRNA reaches the axon: tau mRNA is translated on the site of localization [34] and, in order to be translated in situ, tau mRNA has to be transported to the axon for which HuD protein binds to the uracil-rich 3’UTR in tau mRNA, serving as an anchor protein to kinesin and stabilizing the mRNA molecule so that translation can take place when required within the relatively long period of tau mRNA half-life.
We also investigated other messenger molecules that might localize to the axon. Using bioinformatics tools, we identified a number of mRNAs with uracil-rich 3’UTRs very similar to that in tau mRNA that could also be found in the axon [35,36].
Our findings on tau localization in the neuronal axon together with the protein complex involved in the in situ translation [34] made us the first to demonstrate mRNA axonal stabilization, transport, localization and in situ translation [37].
The implications of tau localization
Tau protein participates in the maintenance of neuronal polarity [13] and axonal transport by binding to microtubules, providing stability and shaping axonal morphology. When tau expression is inhibited, the axon retracts [27]. Efficient axonal transport of synaptic vesicles and organelles such as mitochondria allows the correct functioning of synaptic processes. It has also been shown that tau mRNA was co-localized in the distal part of the axon and growth cone, with the elongation factor 1a, which is a component of the translation machinery [38].
A number of studies have demonstrated that hyperphosphorylation of a truncated form of tau (pTau) prevents microtubule assembly [39] and localizes tau to the cell soma and degenerated neurites [40]. Upon tau hyperphosphorylation, the axon retracts and synapses are lost [41]. Mislocalization of pTau not only causes loss of synapses [42,43] but the accumulation of oligomers in dendritic spines which results in decreased long-term potentiation following internalization of AMPA receptors [42,44], thus affecting neuronal plasticity [45] (Table).
Tauopathies
Reference
Alzheimer Disease
[46]
Argyrophilic grain dementia
[47]
Corticobasal degeneration
[48]
Creutzfeldt-Jacob disease
[49]
Dementia pugilistica
[50]
Down´s syndrome
[51]
Frontotemporal dementia
[52]
Myotonic dystrophy
[53]
Niemann Pick disease, type C
[54]
Pick disease
[55]
Postencephalitic parkinsonism
[56]
Progressive supranuclear palsy
[57]
Table 1: Abnormal localization and hyperphosphorylation of tau, can lead to a spectrum of diseases known as tauopathies.
Conclusion
The demonstration of tau mRNA axonal localization broke with the myth that translation is a process exclusively taking place in the neuronal soma. Tau mRNA transport is essential for efficient neuronal function and maintenance of cell polarity. When tau mRNA is not properly localized to the axon, inefficient neurotransmission at the dendrites and axonal protrusion can be observed, leading to interruptions in transmission of the nerve impulses affecting presynaptic as well as post-synaptic neurons.
References
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996; 65: 693-739.
- Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995; 15: 2219-2230.
- Roy N, Laflamme G, Raymond V. 5' untranslated sequences modulate rapid mRNA degradation mediated by 3' AU-rich element in v-/c-fos recombinants. Nucleic Acids Res. 1992; 20: 5753-5762.
- Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997; 16: 2130-2139.
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009; 29: 4697-4707.
- Yoon BC, Zivraj KH, Holt CE. Local translation and mRNA trafficking in axon pathfinding. Results Probl Cell Differ. 2009; 48: 269-288.
- Uhl GR, Zingg HH, Habener JF. Vasopressin mRNA in situ hybridization: localization and regulation studied with oligonucleotide cDNA probes in normal and Brattleboro rat hypothalamus. Proc Natl Acad Sci U S A. 1985; 82: 5555-5559.
- Wilcox JN, Roberts JL, Chronwall BM, Bishop JF, O'Donohue T. Localization of proopiomelanocortin mRNA in functional subsets of neurons defined by their axonal projections. J Neurosci Res. 1986; 16: 89-96.
- Harlan RE, Shivers BD, Romano GJ, Howells RD, Pfaff DW. Localization of preproenkephalin mRNA in the rat brain and spinal cord by in situ hybridization. J Comp Neurol. 1987; 258: 159-184.
- Trembleau A, Fevre-Montange M, Calas A. [Ultrastructural localization of mRNA coding for oxytocin by in situ hybridization. Study by high resolution autoradiography using a tritiated oligonucleotide probe]. C R Acad Sci III. 1988; 307: 869-874.
- Takemura R, Kanai Y, Hirokawa N. In situ localization of tau mRNA in developing rat brain. Neuroscience. 1991; 44: 393-407.
- Goedert M, Crowther RA, Garner CC. Molecular characterization of microtubule-associated proteins tau and MAP2. Trends Neurosci. 1991; 14: 193-199.
- Litman P, Barg J, Rindzoonski L, Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell culture: implications for neuronal polarity. Neuron. 1993; 10: 627-638.
- Litman P, Barg J, Ginzburg I. Microtubules are involved in the localization of tau mRNA in primary neuronal cell cultures. Neuron. 1994; 13: 1463-1474.
- Drubin DG, Kirschner MW. Tau protein function in living cells. J Cell Biol. 1986; 103: 2739-2746.
- Aronov S, Marx R, Ginzburg I. Identification of 3'UTR region implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999; 12: 131-145.
- Behar L, Marx R, Sadot E, Barg J, Ginzburg I. cis-acting signals and trans-acting proteins are involved in tau mRNA targeting into neurites of differentiating neuronal cells. Int J Dev Neurosci. 1995; 13: 113-127.
- Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997; 124: 3449-3460.
- Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci. 1999; 19: 8818-8829.
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, et al. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991; 67: 325-333.
- Weston JA. Sequential segregation and fate of developmentally restricted intermediate cell populations in the neural crest lineage. Curr Top Dev Biol. 1991; 25: 133-153.
- Shaw G, Kamen R. A conserved AU sequence from the 3’ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986; 46: 659-667.
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994; 265: 615-621.
- Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991; 16: 214-220.
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976; 73: 2424-2428.
- McBurney MW, Reuhl KR, Ally AI, Nasipuri S, Bell JC, Craig J. Differentiation and maturation of embryonal carcinoma-derived neurons in cell culture. J Neurosci. 1988; 8: 1063-1073.
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J Neurosci. 1999; 19: 6907-6917.
- Parnas D, Linial M. Cholinergic properties of neurons differentiated from an embryonal carcinoma cell-line (P19). Int J Dev Neurosci. 1995; 13: 767-781.
- Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996; 271: 11518-11524.
- Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J Neurosci. 2001; 21: 6577-6587.
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985; 42: 39-50.
- Hirokawa N. Mechanism of axonal transport. Identification of new molecular motors and regulations of transports. Neurosci Res. 1993; 18: 1-9.
- Hirokawa N. Axonal transport and the cytoskeleton. Curr Opin Neurobiol. 1993; 3: 724-731.
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002; 115: 3817-3827.
- Aranda-Abreu GE, Hernández ME, Soto A, Manzo J. Possible Cis-acting signal that could be involved in the localization of different mRNAs in neuronal axons. Theor Biol Med Model. 2005; 2: 33.
- Willis D, Li KW, Zheng JQ, Chang JH, Smit AB, Kelly T, et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005; 25: 778-791.
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005; 21: 223-245.
- Malmqvist T, Anthony K, Gallo JM. Tau mRNA is present in axonal RNA granules and is associated with elongation factor 1A. Brain Res. 2014; 1584: 22-27.
- Novák M. Truncated tau protein as a new marker for Alzheimer's disease. Acta Virol. 1994; 38: 173-189.
- Su JH, Cummings BJ, Cotman CW. Early phosphorylation of tau in Alzheimer's disease occurs at Ser-202 and is preferentially located within neurites. Neuroreport. 1994; 5: 2358-2362.
- Takahashi RH, Capetillo-Zarate E, Lin MT, Milner TA, Gouras GK. Co-occurrence of Alzheimer's disease ÃÝ-amyloid and Ï„ pathologies at synapses. Neurobiol Aging. 2010; 31: 1145-1152.
- Miller EC, Teravskis PJ, Dummer BW, Zhao X, Huganir RL, Liao D. Tau phosphorylation and tau mislocalization mediate soluble Aβ oligomer-induced AMPA glutamate receptor signaling deficits. Eur J Neurosci. 2014; 39: 1214-1224.
- Merino-Serrais P, Benavides-Piccione R, Blazquez-Llorca L, Kastanauskaite A, Rábano A, Avila J, DeFelipe J. The influence of phospho-t on dendritic spines of cortical pyramidal neurons in patients with Alzheimer's disease. Brain. 2013; 136: 1913-1928.
- Spires-Jones TL, Hyman BT. The intersection of amyloid beta and tau at synapses in Alzheimer's disease. Neuron. 2014; 82: 756-771.
- Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, et al. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J Neurosci. 2011; 31: 2511-2525.
- Zempel H, Mandelkow E. Lost after translation: missorting of Tau protein and consequences for Alzheimer disease. Trends Neurosci. 2014;.
- Rábano A, Rodal I, Cuadros R, Calero M, Hernández F,Ávila J. Argyrophilic grain pathology as a natural model of tau propagation. J Alzheimers Dis. 2014; 40 Suppl 1: S123-133.
- Chahine LM, Rebeiz T, Rebeiz JJ, Grossman M, Gross RG. Corticobasal syndrome: Five new things. Neurol Clin Pract. 2014; 4: 304-312.
- Krvavica A, Morovic M, Mrden A, Mislov D, Duka-Glavor K, Ivanac K, et al. Alzheimer and Lewy body pathology or Creutzfeldt-Jakob disease. Coll Antropol. 2014; 38: 701-704.
- McKee AC, Cantu RC, Nowinski CJ, Hedley-Whyte ET, Gavett BE, Budson AE, et al. Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J Neuropathol Exp Neurol. 2009; 68: 709-735.
- Mondragón-Rodríguez S, Perry G, Luna-Muñoz J, Acevedo-Aquino MC, Williams S. Phosphorylation of tau protein at sites Ser(396-404) is one of the earliest events in Alzheimer's disease and Down syndrome. Neuropathol Appl Neurobiol. 2014; 40: 121-135.
- Laforce R Jr. Behavioral and language variants of frontotemporal dementia: a review of key symptoms. Clin Neurol Neurosurg. 2013; 115: 2405-2410.
- Caillet-Boudin ML, Fernandez-Gomez FJ, Tran H, Dhaenens CM, Buee L, Sergeant N. Brain pathology in myotonic dystrophy: when tauopathy meets spliceopathy and RNAopathy. Front Mol Neurosci. 2014; 6: 57.
- Zhang M, Wang X, Jiang F, Wang W, Vincent I, Bu B. Mitotic epitopes are incorporated into age-dependent neurofibrillary tangles in Niemann-Pick disease type C. Brain Pathol. 2010; 20: 367-377.
- Kertesz A. The overlapping syndromes of the pick complex. Curr Alzheimer Res. 2011; 8: 224-228.
- Jellinger KA. Absence of alpha-synuclein pathology in postencephalitic parkinsonism. Acta Neuropathol. 2009; 118: 371-379.
- Armstrong RA, Cairns NJ. Spatial patterns of the tau pathology in progressive supranuclear palsy. Neurol Sci. 2013; 34: 337-344.