Abstract
Adolescence covers 10-19 years of human age and is the transition period of life. The prevalence of Iron (Fe) deficiency in Nepal is around 23% population. High Fe content biofortied lines had developed to overwhelm the problem. This study was aimed to explore the effect of Genotype (G) and Genotype × Environment Interaction (GEI) on yield of 16 high Fe content lentil genotypes under four different environments (Nepalgunj, Banke, Dashrathpur, Surkhet, Parwanipur, Bara and Itahari, Sunsari). The research was conducted at the winter season cropping period of the year 2014 and 2015 following randomized complete block design with three replications in each location. Combined analysis of variance showed that there were highly significant differences in genotype and location but not genotype x environment interaction at 5% probability level, where G and GEI captured totally 87.4% of total variability. These four environments had good discriminative and representativeness for yield trait of high Fe content lines. Mean yield performances indicated that biofortifed lentil genotypes HUL-57, PL-4, RL-11 and LG-12 were the best performers across the environments over the years and stability of genotypes indicated that they were satisfactorily stable with high yield.
Keywords: AMMI Biplot and triplot analysis; GEI; Stability; Lentil
Introduction
Lentil (Lens culinaris Medikus subsp. culinaris) is a selfpollinating, diploid (2n = 2× = 14) pulse crop with a relatively large genome of 4,063 Mpb [1]. Generally, lentil varieties are distinguished by seed size and color. Lentil seed sizes are grouped into two types: macrosperma and microsperma. In Nepal, it is the main pulse crop accounting for 62.64% in area and 64.35% in production of the total legume area and production [2]. Lentil statistics showed that area, production and productivity had increased by 100.11%, 270.80% and 85.32% in between 1984/85 and 2012/13 respectively. This crop is also recognized as one of the major agricultural product with high export potential by Nepal Trade Integration Strategy. Obviously Nepalese lentils are rich in micronutrients like iron and zinc and have a great role in nutritional security of low-income people. Nepal has retained its position as the world’s sixth largest producer of lentil in 2012 after it logged 0.64 percent rise in production according to the FAO of the United Nations [3]. In fact, it is the largest exported agricultural item with a share of about 2.3% of total national export from Nepal and about 3.2% of the total world export (ITC, 2010). Nepal ranks fifth (3.2%) in lentil export after Canada (69.5%), USA (10.3%), Turkey (7.3%) and Australia (3.8%). The interplay in the effect of genetic and non-genetic on development is termed as Gx E interaction. G x E interactions are of major consequences to the breeder in the process of evolution of improved varieties. So, phenotypes are the mixture of Genotype (G), Environment (E) components and interactions (GxE) between them. It was reported that the prevalence of Fe deficiency anemia was estimated to be 55.1% in children under five, 31% in school age children, 63.5% in pregnant women, and 35% in manual laborer (Directorate of Public Nutrition, 1993). The problem for women and children is more severe because of their physiological need. The prevalence of Fe deficiency in Nepal is around 23% population. Breeding lentil with high micronutrient content dubbed as ‘biofortification’ provides a cost effective and sustainable solution to combat malnutrition. Yield is a quantitative traits that is strongly affected by environment. GxE study on yield trait of the high Fe and Zn rice lines would give information about the yield and stability, so that it could be selected the best genotypes with high yield and stable across environment. Advanced micronutrient Fe rich breeding materials must be evaluated in multi-location and more than one year. Selection based on the yield performances are the two major phases of varietal development and the later one is highly influenced by the locations and years of testing. The main Environmental effects (E) and Genotype by Environment Interaction (GEI) have been reported as the most important sources of variation for the measured yield of crops [4]. For this reason, multi-location trials are conducted throughout the country for major crops every year. Although the measured yield is a combined result of the effects of the genotype (G), E and GE interaction, only G and GxE are relevant to cultivar evaluation and mega environment identification. Typically, E explains mostly (80% or higher) of the total yield variation, while G and GE are usually small [5]. However, effective interpretation and utilization of MET data in making selection decisions remain a major challenge to researchers. Some important concepts such as mega environment, specific adaptation, and stability all originate from the GE interaction. A significant GE interaction for grain yield can reduce the usefulness of subsequent analysis and limit the feasibility of selecting superior cultivars. Development of widely adapted and or location specific adapted genotypes is the goal of all breeding programs. For this purpose, the genotypes are grown in different environments and their yield stability is estimated before giving any recommendations for variety release. A genotype may be considered to be stable if its environment variance is small. Various methods have been introduced to assume cultivar reaction in different situations. Additive Main Effects and Multiplicative Interaction (AMMI) analysis is one of the popular parametric of multivariate methods to predict adaptation and stability of cultivars. The usefulness of the method to be applied to some different crops has been noted by many researchers [6-8]. Zobel et al. proposed the name AMMI first time. The AMMI model is a hybrid model involving both additive and multiplicative components of two way data structure which enabled a breeder to get precise prediction on genotypic potentiality and environmental influences on it. AMMI uses ordinary ANOVA to analyze the main effects (additive part) and Principal Component Analysis (PCA) to analyze the non-additive residual left over by the ANOVA [9]. Purchase et al., 2000 [10], developed a quantitative stability value to rank genotypes through the AMMI model, named the AMMI Stability Value (ASV). During the analysis of cultivars stability they found a significant correlation between the stability measures ASV with Shukla and Wricke (Wi), and Eberhart and Russel (S2d), while Finlay and Wilkinson (b), and Linn and Binns (Pi) showed limited correspondence with any of the other methods [11,12]. The developed ASV was considered to be the most appropriate single method of describing the stability of genotypes. The breeders want to develop and select high yield and high stability lines which is highly desirable but some time, high yield but low stability lines which is desirable for specific selection, low yield and low stability (desirable for special breeding purposes, e.g. drought resistance selection) while low yield but high stability is undesirable and no one wants to select such types.
Materials and Methods
The present experiment material comprised a total of 16 Fe rich genotypes of lentil were selected on the basis of lab analysis report, yield and yield attributing traits from G x E biofortification trial on lentil conducted at four diverse environments i.e. Nepalgunj, Surkhet, Parwanipur, and Itahari during the year 2014-2015 [13]. The geographical location of Parwanipur is at 27°2’ North Latitude, 84°53’ East Longitude and 115 masl elevation. Climate of testing site falls under subtropical region of Nepal. The average maximum temperature ranges from 22.7°C to 34.52°C and minimum temperature ranges from 8.54°C to 25.9°C with an average annual rainfall of 1550 mm. The rainfall distribution is of unimodal type, 84% of the total rainfall is received from June to September. The geographical location of Surkhet is 81°36’ 0E longitudes and 28°36’ 12N latitude at an altitude of 634m. Mean Above Sea Level (MASL). The average maximum temperature is 32.8°C and minimum temperature is 7.9°C with total sum of rainfall of 236.6 mm along with 72-98% Relative Humidity (RH) during cropping season. Likely the geographical location of Itahari is 87°16’ 60 E longitudes and 26° 40’ 0 N latitude at an altitude of 104 m. Mean Above Sea Level (MASL). The geographical location of Nepalgunj is 810 37” E longitudes and 280 06” N latitude at an altitude of 181m. Mean Above Sea Level (MASL). It has subtropical climate with cool winter (5.4°C) and very hot summer (46°C temperature) with 27-94% Relative Humidity (RH). The annual average rainfall is 1000-1500 mm. However, delayed onset and early termination of monsoon rains, is a regular feature, causing occasional failure of annual crops in the region. The soil on station is sandy loam to silty loam, poor in organic carbon and available N2 but medium in available P2O5 and K2O, pH varies from 7.2-7.5. The trials were conducted in a Randomized Complete Block Design (RCBD) with three replications. The plot size was of 4 meter length of four rows (1m wide) of 25cm spacing between rows and continues between plants. Important quantitative traits i.e. Days to 50% Flowering (DF), Days to 90% Maturity (DM), Plant Height (PH) in cm, number of Pods per Plant (PP), number of Seeds per Pod (SP), 100 Seed Weight (SW) in gram, and grain yield metric ton/ha (GY kg ha-1) were recorded. Five plants randomly selected from each plot to take the data of yield attributing traits. Grain yields were recorded on the plot basis and converted to the kg/ha. Fertilizer was applied @ 20:40:20 kg N, P, K /ha. ANOVA and Stability analysis for yield trait was carried out by using the AMMI model (Agricolae) software R x 64-program version 3.3.2.
Results and Discussions
AMMI analysis of variance
Variance analysis showed that the genotype and environment was found highly significant differences for the yield and yield attributing traits except plant height and pod per plant but non-significant differences in genotype x environment interaction indicating distinct features of genotype and environments but not distinct the genotype x environment interactions [14] on phenotypic expression. High estimate of Sum of Square (SS) for all the traits are expressed by environment (Table 1). Yield variation was observed due to the environment effect indicated that the environments were diverse and a major part of variation in grain yield can be resulted from environmental changes. These data had compiled the requirements for biplot analysis. Biplot analysis was conducted and visualized to determine the differences among the environments, to evaluate stable and wide adaptable line, and to evaluate the environments if a certain graph representing the appropriate environment to select genotypes based on yield. The presence of Genotype Environment Interaction (GEI) was clearly demonstrated by the AMMI model, when the interaction was partitioned among the first three Interaction Principal Component Axis (IPCA). All PCA 1, PCA 2 and PCA 3 were non- significant. PC1 contribute 62.3 % variation to the total. PC2 contribute 25.1 % to the total variation and IPC3 contribute 12.6 % variation of the total with Pr. F value more than 0.005. It means that by using PC1 and PC2, the analysis could explain 87.4 % variation (Table 2).
Source of variation
Df
Sum Sq
Mean Sq
F value
Pr(>F)
ENV
3
17440986
5813662
56.4975
9.972e-06 ***
REP(ENV)
8
823210
102901
1.8164
0.07713.
GEN
15
2849535
189969
3.3532
6.042e-05 ***
ENV:GEN
45
2570246
57117
1.0082
0.46809
Error
168
9517638
56653
CV%24.58
Table 1: Combine variance analysis of yield trait of high Fe content lentil in 4 sites.
Principle component
percent
acum
Df
Sum.Sq
Mean.Sq
F.value
Pr.F
PC1
62.3
62.3
17
1292529.7
76031.16
1.34
0.1736
PC2
25.1
87.4
15
521788.9
34785.93
0.61
0.8641
PC3
12.6
100.0
13
260968.4
20074.49
0.35
0.9824
Table 2: Analysis of variance of principle components of biplot genotype and location of the trial of high Fe lentil lines in four sites, 2014 and 2015.
Mean yield and yield attributing traits comparison
The pooled mean yields and yield attributing traits of all the environments are presented in Table 3. AMMI analysis variance for yield and yield attributing traits indicated that significant differences were observed in genotype, environment and Genotype environment interaction in the days to flowering, days to maturity, hundred seed weight and plant height while significant differences was genotype and environment but in Genotype x Environment interaction in the yield trait. Only nine genotypes have produced grain yield above the grand mean yield, while all the checks produced below the grand mean. The highest mean grain yield of genotypes averaged over environments was produced by HUL-57 (1179 kgha-1) followed by PL-4 (1119 kg ha-1) and RL-11 (1077 mt ha-1) while lowest by ILL4605 (610 kg ha- 1). Different genotypes showed in consistent performance across all environments. The highest environments mean grain yieldover genotypes was recorded from Nepalgunj (1311 kg ha-1) followed by Itahari (938 kg ha-1) and surkhet (776 kgha-1), while the lowest mean grain yield (432 kg ha-1) was produced at Parwanipur. The high yielding genotypes HUL-57, PL-4, RL-11 are stable and suitable for all the environments.
SN
Name of entries
DF
DM
Plht
P/P
Grain Yield
(kg/ha)
HSWT
1
ILL-8006
83
134
29
77
963
2
2
RL-6
81
131
30
66
1042
1
3
RL-12
84
133
30
77
1070
2
4
ILL-7715
85
133
31
76
1039
1
5
ILL-7164
85
133
31
76
1029
2
6
ILL-3490
84
134
30
78
884
1
7
Khajura- 2
84
132
30
85
984
2
8
Simal
86
133
29
67
851
1
9
Shital
81
130
30
75
996
1
10
Sagun
86
131
29
68
1013
2
11
HUL-57
85
133
31
76
1179
1
12
LG-12
84
131
30
77
1061
1
13
PL-4
82
133
30
70
1119
2
14
RL-11
84
134
31
74
1077
2
15
RL-4
85
133
29
69
912
2
16
ILL-4605
68
126
25
55
610
3
Mean
83
132
30
73
989
2
F-pr
Genotype
<.001
<.001
0.008
0.189
<.001
<.001
Environment
<.001
<.001
<.001
<.001
<.001
<.001
GxE
<.001
0.01
0.707
0.075
175.1
<.001
CV%
4.9
2.4
13.7
31.2
27.7
16.4
LSD
2.957
2.29
2.919
16.57
175.1
0.1915
Table 3: AMMI analysis of variance for yield and yield attributing traits of lentil genotypes across 4 environments.
The AMMI Stability Value (ASV) and AMMI stable index are calculated as suggested by Zobel et al, 1998 and Purchase et al. [10] and their ranks are presented in Table 4. The highest mean grain yield of genotypes averaged over environments were produced by HUL-57 (1179 kg ha-1) followed by PL-4 (1119Kg ha-1) and RL-11 (1077 mt ha-1) while lowest by ILL4605 (610 kg ha-1). The genotypes which has low stability value (ASV) is said to be stable and the breeder chose the stable genotypes, having grain yield above the mean grand yield. In this experiment genotype HUL-57 ranked 1st instability followed by RL-12, RL-11 and LG-12 and suitable for all environment but out of test genotypes; genotypes HUL-57, RL-12, RL-11, LG-12, Sagun, PLGenotypes 4, RL-6, ILL-7715, ILL-7164 produced the mean yield above grand mean (Table 4).
Genotypes
Mean Yield
(Kg ha-1)
ASV
YSI
rASV
rYSI
HUL-57
1179
13.18
14
13
1
RL-12
1070
1.79
3
1
2
RL-11
1077
3.21
6
3
3
LG-12
1061
8.18
13
9
4
Sagun
1013
5.84
11
6
5
PL-4
1119
16.02
20
14
6
RL-6
1042
6.37
14
7
7
ILL-7715
1039
3.25
12
4
8
Khajura- 2
984
9.63
20
11
9
ILL-7164
1029
16.68
25
15
10
Shital
996
8.91
21
10
11
ILL-8006
963
1.83
14
2
12
RL-4
912
7.47
21
8
13
ILL-3490
884
10.78
26
12
14
Simal
851
5.39
20
5
15
ILL-4605
610
28.13
32
16
16
Table 4: Mean grain yield (kgha-1), AMMI Stability Values (ASV), stability index and ranking orders of the 16 genotypes of lentil.
AMMI 1 biplot
Biplots are graphs where aspects of both genotypes and environments are plotted on the same axes so that inter relationships can be visualized. There are two basic AMMI biplot, the AMMI 1 biplot, where the main effects of grain yield (genotype mean and environment mean) and IPCA1scores for both genotypes and environments are plotted against each other. On the other hand, the second triplot is AMMI 2 where scores for IPCA1 and IPCA2 are plotted.
In the AMMI 1 biplot, the usual interpretation of biplot is that the displacements along the abscissa indicate differences in main (additive) effects, whereas displacements along the ordinate indicate differences in interaction effects. Genotypes that group together have similar adaptation while environments which group together influences the genotypes in the same way [15]. The graph showed that the genotypes which are in the right side of perpendicular i.e sagun, RL-11 and ILL4605 genotypes are less affected by G x E inter action. Genotypes HUL-57, RL-12, RL-11, LG-12 and Sagun were more close to the center point and indicated that stable across the environments (Figure 1 and 2).
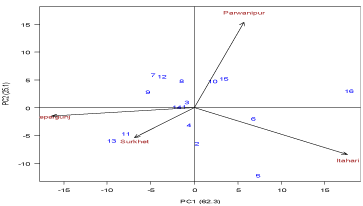
Figure 1: AMMI 1 Biplot for grain yield (kg ha-1) of 16 lentil genotypes and
eight environments using genotypic and environmental scores.
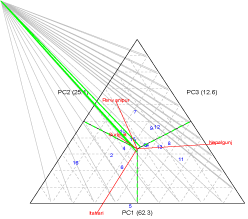
Figure 2: AMMI 1 triplot for grain yield (kg ha-1) of 16 lentil genotypes and
eight environments using genotypic and environmental scores.
AMMI 2 triplot
The environmental scores are joined to the origin by side lines. Sites with short arrow do not exert strong interactive forces. Those with long arrow exert strong interaction. The genotypes close to ordinate expressed general adaptation, whereas the further genotypes depicted more specific adaptation to environments [16,17]. All the environments Nepalgunj, Surkhet, Parwanipur and Itahari are connected to the origin. The environments Surkhet and Parwanipur had short spokes and they do not exert strong interactive forces. The genotypes occurring close together on the plot will tend to have similar yields in all environments, while genotypes far apart may either differ in mean yield or show a different pattern of response over the environments. Hence, the genotypes near the origin are not sensitive to environmental interaction and those distant from the origins are sensitive and have large interaction. In the present study genotype PL-4, simal (Nepalgunj), ILL3490 (Itahari), Sagun (Surkhet) and, RL-4 and Khajura-2(Parwanipur) are more responsive to the environment given in parenthesis and are specific adopted. The genotypes ILL-8006, RL-11, and RL-12 are less affected by the G x E interaction and thus would perform well across a wide range of environments.
Conclusion
Yield is a quantitative trait that is strongly affected by environment. AMMI statistical model might be a great tool to select the most suitable and stable high yielding genotypes for specific as well as for diverse environments. In the present study, AMMI model has shown that the largest proportion of the total variation in grain yield was attributed to environments. The genotype HUL-57, PL-4, RL-11 and LG-12 showed higher grain yield than all other genotypes across all the environments and performed well at all the environment. The genotypes ILL-8006, RL-11, and RL-12 are less affected by the G x E interaction and thus would perform well across a wide range of environments.
Acknowledgment
The research was supported collaboratively by ICARDA through Harvest plus Biofortification Project on Development of Lentil Cultivars with High Concentration of Iron and Zinc for Nutritional Security. We are highly acknowledged to Dr. A. Sarker, Regional Coordinator, ICARDA and China Regional program for bringing such a wonderful project. We would like to give special thanks to NARC ED Dr. YR Pandey, CHD Dr. YP Giri and Planning Director Dr. AK Gautam for his continuing support to the program. Most importantly we would like to thanks all the staffs that directly and indirectly help to conduct the experiments.
References
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Boil. 1991; 9: 208-218.
- Statistical Information on Nepalese Agriculture 2013/14. Ministry of Agriculture and Development business promotion and agriculture statistics division. Nepal. MOAD. 2014.
- FAOSTAT. Statistical Database. FAO. 2010.
- Dehghani H, Ebadi A, Yousefi A. Biplot analysis of genotype by environment interaction for barley yield in Iran. Agron J. 2006; 98: 388-393.
- Yan W, Kang MS. GGE-biplot analysis: Agraphical tool for breeders, geneticists, and agronomists. CRC Press, USA. 2003.
- Abay F, Bjornstad A. Specific adaptation of Barley varieties in different locations in Ethiopia. Euphytica. 2009; 167: 181-195.
- Alwala S, Kwolek T, McPherson M, Pellow J, Meyer D. A comprehensive comparison between Eberhart and Russell joint regression and GGE biplot analyses to identify stable and highyielding maize hybrids. Field Crops Res. 2010; 119: 225-230.
- Annicchiarico P, Harzic N, Carroni AM. Adaptation, diversity, and exploitation of global white lupin (Lupinus albus L.) landrace genetic resources. Field Crops Res. 2010; 119: 114-124.
- Gauch HG. AMMI and related analysis for two way data matrics. Microcomputer power, Ithaca, New York. USA J. 1993; 80: 388-339.
- Purchase JL, Hatting H and Van Deventer CS. Genotype x environment interaction of winter wheat (T.aestivum) in South Africa: Stability analysis of yield performance. S Afr J Plant Soil. 2000; 17: 101-107.
- Eberhart SA, Russell WA. Stability parameters for comparing varieties. Crop Sci. 1966; 6: 36-40.
- Finlay KW, Wilkinson GN. The analysis of adaptation in a plant breeding programme. Aust j Agric Res. 1963; 14: 742-754.
- Yadav NK, Ghimire SK, Sah BP, Sarker A, Shrestha SM, Sah SK. Genotype x environment interaction and stability analysis in lentil (Lens culinaris Medik.). Paper published in the International Journal of Environment, Agriculture and Biotechnology (IJEAB). 2016; 1: 2456-1878.
- Freeman GH. Statistical methods for the analysis of Genotype X Environment Interactions. Heredity. 1973; 31: 339-354.
- Kempton RA. The use of biplots in interpreting variety by environment interactions. Journal of Agricultural Science. 1984; 103: 123-135.
- Ebdon J, Gauch H. Additive main effect and multiplicative interaction analysis of national turfgrass performance trials: I. Interpretation of genotype X Environment Interaction. Crop Sci. 2002; 42: 489-496.
- Gauch HG, Zobel RW, Kang MS & Gauch HG. AMMI analysis of yield trials. Genotype by-environment interaction, pp: 85-122. Gujarat Agric Uni Res J. 1996; 22: 101-102.
