
Review Article
Austin J Anal Pharm Chem. 2016; 3(1): 1058.
An Analytical and Computational Infrared Spectroscopic Review of Vibrational Modes in Nucleic Acids
A Heidari*
Faculty of Chemistry, California South University, 14731 Comet St. Irvine, CA 92604, USA
*Corresponding author: A Heidari, Faculty of Chemistry, California South University (CSU), 14731 Comet St. Irvine, CA 92604, USA
Received: February 16, 2016; Accepted: March 07, 2015; Published: March 09, 2016
Abstract
In this review, we are reviewing single nucleotide bases using Attenuated Total Reflectance (ATR–FTIR) spectra before moving onto more complex oligionucleotides and ultimately DNA/RNA. This work will lay a foundation to understanding the spectroscopy of DNA/RNA in response to hydration and has important implications for understanding how cells respond to environmental stresses such as dehydration. Also, we are using quantum chemical methods to model DNA/RNA conformation to predict the frequencies and intensity differences in ATR–FTIR spectra. The computational review will initially focus on modelling small– to medium–sized oligonucleotides with the view of correlating predicted results with experimental spectra of these oligonucleotides. The effect of explicit and implicit solvent will be reviewed concurrently.
Keywords: Nucleic Acids; Vibrational Modes; Infrared Spectroscopy; Quantum Chemical Methods; DNA/RNA Conformation; ATR–FTIR Spectra; Modelling; Analytical Chemistry; Computational Chemistry
Background
Deoxyribonucleic acid (DNA) is the fundamental molecule of life encoding the genetic code for the development and functioning of every living organism and a large variety of viruses. RNA, proteins, and DNA are the main macromolecules, which are necessary for every form of life. The genetic system is encoded in the form of a sequence of nucleotides (guanine, adenine, thymine, and cytosine), which are denoted by the letters G, A, T, and C. Most DNA molecules consist of double–stranded helices (Figure 1) [1], composed of two long polymers made of simple units named nucleotides, molecules with backbones constructed of alternating sugars (deoxyribose) and phosphate groups (relative of phosphoric acid), with the nucleobases (G, A, T, C) joined to the sugars. DNA is a suitable store for biological information, because the DNA backbone resists cleavage and the double–stranded structure allows the molecule to have a built–in duplicate of the encoded data.
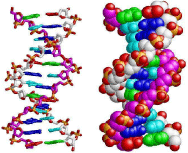
Figure 1: Modelling of section of a DNA double helix [1].
In DNA, the two strands are located in opposite directions to each other. Consequently, these strands are anti–parallel; one backbone is 3’ (three prime) and the other 5’ (five prime). This presents the direction of the 3rd and the 5th carbon, which is facing on the sugar molecule. One of four types of molecules named nucleobases is attached to each sugar (bases). Data is encoded based on the sequence of these four nucleobases placed along the backbone. The genetic code is then used to read this information. In fact the genetic code tries to determine the sequence of the amino acids inside proteins [2]. The code is read by transcribing extensions of DNA into the related nucleic acid RNA.
DNA is structured in chromosomes as long structures inside cells. In the process of cell division, these chromosomes are duplicated in a process called DNA replication. This process fully furnishes each cell with its full–required set of chromosomes. In eukaryotic organisms (animals, plants, fungi, and protists), most of DNA is stored inside the cell nucleus and partly inside organelles; the examples of the latter are mitochondria or chloroplasts [3]. On the contrary, in prokaryotes (i.e., bacteria), DNA is stored just in the cytoplasm. It should be added that compaction and management of DNA is occurred inside the chromosomes by chromatin proteins such as histones. The interactions between DNA and other proteins are guided by these compact structures, which assist in controlling which parts of the DNA are copied (Figures 2 & 3).
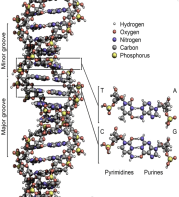
Figure 2: The schematic structure of DNA double helix. Different atoms in the
structure are presented using color codes. Lower right is detailed structure of
two base pairs [2].
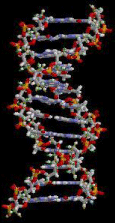
Figure 3: The schematic structure of section of a DNA double helix [2].
History of DNA Research
The Swiss physician, Friedrich Miescher, was the first one who identified DNA in 1869. He discovered microscopic matter in the pus of disposed surgery bandages. He named it “nuclein” due to the fact that it was located inside the cell nuclei [4]. Later on in 1878, Albrecht Kossel, identified the non–protein part of “nuclein” (i.e. nucleic acid) and its five primary nucleobases [5]. In 1919, Phoebus Levene, discovered the base, sugar and phosphate nucleotide unit [6]. Levene believed that DNA comprised of a chain of nucleotide units, which are connected together by the phosphate groups. It should be added that Levene believed that the string was short and the repetition of bases occurred in a fixed state. It was William Astbury who created the first X–ray diffraction patterns in 1937. These patterns showed that DNA is regularly structured [7].
Nikolai Koltsov suggested in 1927 that descended traits were descended through a “giant hereditary molecule” constituted of “two mirror strands that would replicate in a semi–conservative fashion using each strand as a template” [8, 9]. Avery et al. were first to develop a system that clearly showed that DNA held genetic data— the Avery–MacLeod–McCarty experiment. They found in 1943 that DNA was the storage vector for genetic data [10]. It was in 1952 that the role of DNA in heredity was proved by Alfred Hershey and Martha Chase. The Hershey–Chase experiment demonstrated that DNA was the genetic material of the T2 phage [11].
In the journal “Nature” in 1953, James D. Watson and Francis Crick proposed what was later known as the first correct double–helix model of DNA structure [12]. At that time, they based their double– helix, molecular model of DNA on a single X–ray diffraction image (named “Photo 51”) [13], taken in May 1952 by Rosalind Franklin and Raymond Gosling, and Erwin Chargaff’s information provided to them through private contact in the previous years, suggesting that the DNA has paired bases. Chargaff highly contributed to the understanding of double–helix configurations of B–DNA and A– DNA (see section 4).
In the same issue of “Nature” five papers were presented, which provided experimental evidence in support of the model proposed by the Watson and Crick [14]. One of these was a research paper by Franklin and Gosling, which presented their X–ray diffraction data, and the original analysis approach that supported the Watson and Crick’s model to some extent [15,16]. This issue also presented a paper on DNA structure by Maurice Wilkins et al. Their analysis and in vivo B–DNA X–ray patterns supported in vivo presence of double– helical DNA configurations, suggested by Crick and Watson for their double–helix molecular model of DNA in another article in the same issue of “Nature” [17].
Watson, Crick, and Wilkins were jointly awarded the Nobel Prize in Physiology and Medicine in 1962, unfortunately after the death of Franklin [18]. At that time Nobel Prizes were not awarded posthumously. To whom the discovery is to be attributed is still the subject of debate [19].
In an important representation in 1957, Crick founded the principal dogma of molecular biology, suggesting how DNA, RNA, and proteins are related, and establishing the “adaptor hypothesis” [20]. A year later in 1958, in an experiment by Meselson–Stahl the final evidence for the replication mechanism as an implication of the double–helical structure was obtained [21]. More studies by Crick et al. affirmed that non–overlapping triplets of bases, called codons, are the basis of the genetic code [21,22]. This enabled Har Gobind Khorana, Robert W. Holley and Marshall Warren Nirenberg to decode the genetic code [22]. All of these findings constitute the formation of a discipline called molecular biology.
Structure
The structure of DNA of every species consists of two helical chains. Each of them is wound around the same axis, and each of has a pitch of 3.4 nm and a radius of 10 ångströms (1.0 nanometers) [12]. Another study suggested that when the DNA chain is measured in a specific approach, its width is between 2.2 to 2.6 nm, and length of a nucleotide unit was 0.33 nm [23]. Each individual repeating unit is very small, but considering the number of nucleotides, it is possible to have DNA polymers made from big molecules. For example, the largest human chromosome, i.e., chromosome 1, is around 220 million base pairs long [24].
In living organisms, there are usually no individual DNA molecules. However there are some paired molecules, which are strongly bound [25,26]. These two long strands are twisted in the same way as vines do to form a double helix shape. The nucleotide repetitions comprise the part of the molecule backbone, binding the chains together, as well as nucleobase that is in interaction with the other DNA strand in the double helix. A nucleobase, which is connected to a sugar, is called a nucleoside, and a base connected to a sugar as well as one or more phosphate groups is known as a nucleotide. In addition, a polynucleotide is a kind of polymer, which contains several nucleotides connected together (e.g., in DNA) [27].
The backbone of the DNA strand comprises alternating residues of phosphate and sugar [28]. The sugar in DNA constitutes a pentose (five–carbon) sugar (i.e., 2–deoxyribose). Phosphate groups link the sugars. Phosphate groups provide the phosphodiester bonds of the carbon atoms number 3 and 5 of sugar rings. The asymmetric nature of bonds demonstrates that a strand of DNA is directional. The nucleotides of one strand and another are oppositely oriented, which means that strands are antiparallel. The asymmetric endpoints of DNA strands are known as 5′ (five prime) and 3′ (three prime) ends. The 5’ and 3’ ends have a terminal phosphate group and a terminal hydroxyl group, respectively. Sugar constitutes one of the principal differences between DNA and RNA, as RNA has pentose sugar ribose in place of DNA’s 2–deoxyribose [26] (Figure 4).
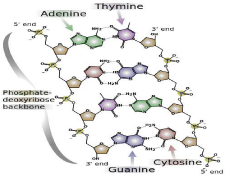
Figure 4: DNA’s chemical structure. Dotted lines denote hydrogen bonds [2].
Two forces are mainly responsible for stabilization of DNA double helix: the first force is provided by hydrogen bonds of nucleotides and the second force is provided by base–stacking interactions between aromatic nucleobases [29]. In the aqueous environment of the cell, the conjugated π bonds of nucleotide bases are arranged in a line, which stands perpendicular to the axis of the DNA molecule. This fact causes the interaction with the solvation shell to be minimal and thus the Gibbs free energy will be minimized. A full nucleotide mainly consists of these bases, which are attached to sugar/phosphate. The following figure illustrates the bases for adenosine monophosphate (Figure 5).
Figure 5: A section of DNA. The bases stand between two spiral strands in a
horizontal direction (Schematic view) [30].
Other DNA Structures
There are many possible forms of DNA, such as A–DNA, B–DNA, and Z–DNA forms. However, B– DNA and Z–DNA are the only ones that have been directly found in functional organisms [28]. The form DNA takes is determined by the level and direction of super coiling, DNA sequence, the hydration level, chemical modifications of the bases, the concentration and the kind of metal ions, and whether or not polyamines exist in solution [30,31].
The first published papers on A–DNA X–ray diffraction patterns, and also B–DNA based their analyses on Patterson transforms. These papers presented little data on the structure of directional fibers of DNA [31, 32]. Later in 1953, Wilkins et al. suggested another analysis for in vivo B–DNA X– ray diffraction/scattering patterns of highly hydrated DNA fibers using squares of Bessel functions [32]. James D. Watson and Francis Crick published in the same journal their paper on molecular modelling analysis of the DNA X–ray diffraction patterns, and showed it was the double–helix structure [12].
The “B–DNA form” is the most common form occurring in cells conditions [33], yet it is not a family of related DNA conformations rather than a well–defined conformation [34], which is found when hydration levels are high, which is typical of living cells. Their X–ray diffraction and scattering patterns characterize molecular paracrystals that have a significant level of disorder [35,36].
The A–DNA form is a right–handed spiral, which is wider than B–DNA. It has a wide and shallow minor groove, while its major groove is relatively narrower and deeper. The A form is found in non– physiological conditions present in partly dehydrated DNA samples. However, under the cell conditions, it can be formed in hybrid pairings of DNA and RNA strands, and also in enzyme–DNA complexes [37,38]. It may be possible that conformation of those parts of DNA in which bases have chemically modified by methylation change more, and become shaped like Z. The strands create a left– handed spiral around helical axis, which is the opposite of more frequently found B form [39]. These rate structures are identifiable using specific Z–DNA binding proteins, and play a role in regulation of transcription [40] (Figure 6).
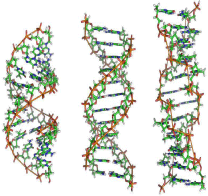
Figure 6: From left to right, the structures of A, B, and Z DNA [41].
General Characteristics of FT–IR Spectra of Nucleic Acids
Vibrations of nucleic acid are caused by various segments of the macromolecule, and the corresponding IR absorptions are found in several spectral regions. The major vibrations in the regions of spectrum ranges from 1800 cm-1 and 700 cm-1, and four distinct different areas are identified: from 1800 cm-1 to 1500 cm-1 absorption bands because of the stretching vibrations of double bonds in the base planes; from 1500 cm-1 to 1250 cm-1 bands because of the base– sugar entities, which specifically highly rely on the glycosidic torsion angle; from 1250 cm-1 to 1000 cm-1, because of high absorptions of the phosphate groups and the sugar; and less than 1000 cm-1 bands because of the vibrations of phosphodiester chain paired with sugar vibrations.
In the case of exposure of a DNA film to an atmosphere in which hydration is controlled by saturated solutions of known salts, the share of water will have lower importance, compared with an aqueous solution. If we consider the data carefully, we could identify an infrared spectrum even in the 1800 cm-1 to 1500 cm-1 range. In case of exposure of the film to vaporized D2O, an exchange of H2O and D2O results, which can be identified in the IR spectrum? Key alterations of the DNA spectrum showing the exchange of labile hydrogens of the bases, specifically the NH2 groups of adenines, guanines, the NH groups of thymines and cytosines are observed. A slow exchange of hydrogens bound to C–8 of the purine base moieties is only observable at high temperatures.
Solid–state analysis of DNA with FTIR microscopy is possible using micro–crystals. IR spectroscopy– specific microscopes with mirrors that wholly reflect CASs grain type optics are currently found in the market, which can be used along with FT–IR spectrophotometers.
Characterization of Nucleic Acid Secondary Structures
In solutions with weak ionic strength, native DNAs and most artificial two–stranded DNAs, multiple and oligonucleotides acquire a B–type conformation. In identical circumstances, two–strand RNAs acquire A–form conformation. In particular the phosphate antisymmetric stretching vibration occurs at 1226 cm-1 in the B form and at 1245 cm-1 in the A form, and a band which shows the base pairing of the nucleic acid is moved from 1716 to 1712 cm-1.
The A–type conformation of DNA is also seen in DNAs as the water content varies in the nucleic acid sample. Reducing the relative humidity in the environment of DNA film is a simple way to fulfill this end. If relative humidity (R.H.) is reduced from 100% R.H. to 58% R.H., conformation of some DNA sequences will be shifted from B to A. The double–helix structure will be disorganized as hydration is reduced even more. This shift does not occur in some DNA forms.
The two A– type and B–type conformations of DNA may also be described according to band pattern which shows the vibrations of the bases along with vibrations of the sugar, which band is placed in 1250–1400 cm-1 range.
DNA conformations may also be characterized by polarized infrared radiation. If the sample has a special orientation axis, for example, in stretched fibers or films, the infrared spectra formed by incident radiation will be different, whose electric field E has perpendicular or parallel polarization with respect to the direction of the molecule axis. Computation of directions of transition dipole moments in the molecule can be conducted using these dichroic spectra, and in simple cases, they provide data about geometric parameters [42,43]. It is possible to conduct these computations regarding absorption bands of the antisymmetric and symmetric movements of the phosphate groups [44]. As mentioned in the introduction, it is possible to apply these IR bands, which are sensitive to conformation to calculate the two A and B helical geometries if the sample contains both of them.
Absorption Band Attributes
Both RNA and DNA have a broad intense band at 3400 cm-1 (or at 2500 cm-1 if deuterated), which is attributed to the OH (or OD) stretching vibration of absorbed water molecules. In the range from 1800 to 1500 cm-1, strong bands are located because of the C=O stretching, skeletal stretching, and NH bending vibrations of the organic base residues [45]. The PO2¯ antisymmetric stretching vibration occurs at 1220 cm-1 for B–DNA and ~1240 cm-1 for A–DNA [45,46]. A number of intense bands located in the 1100–1000 cm-1 region are attributed to the PO2¯ symmetric stretching vibration of the phosphate group, and the C–O stretching vibrations of the ribose or deoxyribose moiety [45–47]. The region ranging from 1000 cm-1 to 700 cm-1 has a number of medium or weak bands, derived from C–O stretching, the P–O stretching, and NH out–of–plane bending vibrations and adsorbed water molecule vibrations. The region ranging from 600 cm-1 and 400 cm-1covers the D2O molecules and ND out–of– plane vibrations.
DNA Analysis
Infrared spectroscopy can be used to analyze ribonucleic acid (RNA), deoxyribonucleic acid (DNA), and nucleic acids [48–52]. The division of nucleic acids spectra into different states is possible according to its components, that is, its base, sugar, and phosphate groups. The bases (guanine, uracil, thymine, adenine, and cytosine) cause pyrimidinic and purinic vibrations ranging 1800–1500 cm−1, with these bands constituting sensitive markers for base stacking and base pairing effects. Nucleic acids bands ranging 1500–1250 cm−1 arise from vibrational pairing of a sugar and a base, and we see sugar– phosphate chain vibrations in the region ranging 1250–1000 cm−1. Sugar/sugar–phosphate vibrations are seen in ranges from 1000 to 800 cm−1. Table 1 provides a list of the main infrared states of nucleic acids [53] (Table 1).
Wave number (cm-1)
Assignment
2960–2850
CH2 stretching
1705–1690
RNA C=O stretching
1660–1655
DNA C=O stretching, N–H bending RNA C=O
stretching
1610
C=C imidazole ring stretching
1578
C=N imidazole ring stretching
1244
RNA PO2– asymmetric stretching
1230
DNA PO2– asymmetric stretching
1218
RNA C–H ring bending
1160, 1120
RNA ribose C–O stretching
1089
DNA PO2– symmetric stretching
1084
RNA PO2– symmetric stretching
1060, 1050
RNA and DNA Ribose C–O stretching
1038
RNA ribose C–O stretching
1015
DNA ribose C–O stretching
RNA ribose C–O stretching
996
RNA uracil ring stretching, uracil ring bending
970, 916
DNA ribose–phosphate skeletal motions
Table 1: Skeletal motions of DNA ribose–phosphate [53].
In case of structural analyses, recording of infrared spectra of nucleic acid fibers requires that they are kept in an environment whose relative humidity is known, because it is established that crystallinity changes as the humidity of the air in the environment of the fibers changes. Such conditions are achieved if one chooses the directional nucleic acid film located on a plate of BaF2, CaF2 or AgCl, and seals it with a second plate and a space above a chamber that contains a saturated solution of the needed salt [45]. Spencer [54] presented solutions, which provide the correct constant humidities. By vapour–phase exchange with a D2O solution of the right salt in the chamber of the sealed cell, which was presented above, it will be possible to virtually deuterate a nucleic acid film. Placing the nucleic acid film in the cell and a D2O salt solution at the bottom of the cell causes almost complete replacement of the hydrogen atoms in the film by deuterium atoms, except for CH hydrogens.
Sutherland and Tsuboi [45], who analyzed the polarized spectra of directional films of sodium DNA at 12 varying humidities, stated that there is a correlation between their findings and structure provided by Crick and Watson [55] by Feughelman et al. [56]. Bradbury et al. provided an example of this kind of analysis of film [57].
Bradbury et al. [57] provided a detailed description of the experimental methods applied in film analysis of DNA polymers, and presented polarized infrared spectra of directional sheets of DNA lithium and sodium salts, in deuterated and undeuterated modes in humidities ranging from 0% to 94% R.H. Two types of sodium DNA were found by them, one form at humidities above 90% R.H.,at which bases are perpendicular to the helical axis (B–type), and another at humidities from 70 to 80% R.H., at which the bases have bent by an angle lower than 13° to the normal to the helical axis (A– type). At humidities above 66% R.H., the B–form of Lithium DNA is found, and in another, different from the A–form of sodium DNA is found at humidities from 44 to 56% R.H. at which humidities the bases are bent around 4° from the vertical (C–type). These authors addressed direction of the phosphate groups, as a conclusion from dichroic effects.
Computational Study
Computational quantum chemistry solves the Schrödinger equation for molecular systems. It is a branch of chemistry that intercalates with physics and mathematics and is based on principles of quantum mechanics. The methods of computational quantum chemistry have been coded in a number of quantum chemical packages to calculate molecular properties as well as kinetics and thermodynamics of chemical reactions. Due to its predictive power methods of computational quantum chemistry have become complementary to experiment over the last decade and as a result, these methods have been widely used for designing new materials and drugs.
Equilibrium structures, interaction energies, chemical reactivity, distributions of charge, dipoles and higher multipole moments, and other spectroscopic quantities such as vibrational frequencies are among molecular properties that can be predicted by means of computational chemistry methods.
Computational chemistry methods are divided into two groups: (1) wave function–based (WF) such as methods of Møller–Plesset perturbation theory and coupled cluster theory and (2) electron density– based methods such as density functional theory (DFT). The wave function–based methods scale exponentially with molecular size and therefore their applications to large molecular systems have been limited. Although DFT–based methods scale linearly with molecular size, they have limitations in treatment of non–covalent interactions, especially π–π stacking and van der Waals interactions [58– 61].
The theoretical studies performed so far have focused on understanding (1) energetics of specific interactions such as hydrogen bonding and π–π stacking in complexes of nucleobases and (2) effect of theses specific interactions on vibrational frequencies.
Energetics of Specific Interactions such as Hydrogen Bonding and π–π Stacking in Complexes of Nucleobases
Non–covalent interactions determine the structure of large biomacromolecules such as DNA, RNA, and proteins, thus forming the necessary building blocks. These blocks can be neutral or charged, with various intermolecular forces being responsible for their stabilization. The dominating forces are predominantly electrostatic hydrogen (H–) bonding and purely Coulomb (electrostatic) interactions [62]. Hydrogen bonding in DNA bases is important in providing stability of the DNA double helix and the specificity for information transfer. Other fundamental forces include dispersion, induction, and charge–transfer. The strength of stacking interactions was considered to be much weaker than the H– bonding for a long time as evidenced in noble gases. Recent calculations showed that stabilization energies of stacked DNA base pairs could in fact be very large, almost equating that of H–bonding. The stacking interactions were originally defined as through–space interactions between aromatic systems [63]. Nowadays, the stacking takes on a more general definition and is applied to systems with delocalized electrons, thus playing a much wider role in biology overall.
It is difficult to determine the role of fundamental energy contributions in extended biomacromolecules, as the applications of accurate quantum chemical methods have been limited. Therefore, a biomacromolecule is usually broken into smaller fragments (e.g. DNA nucelobases) that become tractable at a high quantum chemical level of theory. Model complexes incorporating hydrogen bonding and stacking between these fragments can be easily accessed by more accurate quantum chemical methods. The recent progress in quantum and computational chemistry have made it possible to predict the interaction energies of extended complexes with more than 24 atoms (the size of the benzene dimer) at the CCSD(T) level in the complete basis set limit [64–67]. The CCSD(T) method is highly accurate and is considered the benchmark method for studying intermolecular non–covalent interactions [68]. Due to the progress in computational power a number of papers on the accurate interaction energies of DNA base pairs, H–bonded and stacked DNA and RNA base pairs and amino acid pairs have been published since 2003 [63,69–74]. When selecting a computational methodology for studying these complex interactions particular care needs to be taken with respect to the quality (size) of the basis set and inclusion of electron correlation effects. CCSD(T) is a very expensive method and therefore, a more computationally feasible method is needed to study systems consisting of > 24 atoms. With respect to its much more affordable cost, a second order Møller–Plessett perturbation theory (MP2) is considered an alternative method to CCSD(T) for studying intermolecular interactions. In studying the performance of cheaper methods with the various basis sets the following features need to be considered: (1) overall average performance for a particular basis and (2) quality of the results obtained for each type of interaction. For example, if a method produces excellent results for hydrogen bonding and mixed interaction but describes dispersion interactions only weakly, it might yield a fairly good overall average interaction energy error, but it could not be considered as a good general method for treating non–covalent interactions. To be considered reliable, a computational method needs to provide a balanced description for all types of intermolecular forces. A number of computational studies have been conducted in the last decade to determine the right combination of the level of theory and the basis set that is most suited for accurate description in intermolecular interactions between DNA base pairs. Brief highlights of those studies are presented below.
Interaction energies and geometries for more than 100 DNA base pairs, amino acid pairs and model complexes were calculated using the MP2 level of theory with a number of basis sets and the calculated results were compared with the CCSD(T)/CBS level of theory [75]. MP2 level of theory was insufficient to treat dispersion interactions and therefore, a correction for higher order correlation effects through CCSD(T) must be used, whenever significant dispersion contribution occurs. For complexes with weaker hydrogen bonds as those in amino acid pairs the MP2 level of theory already recovered the majority of correlation effects.
Hydrogen–bonded DNA base pairs were studied using the HF and MP2 levels of theory. These base pairs contained monomers of different polarity and a wide range of secondary long–range electrostatic interactions, including those of hydrogen atoms bonded to ring carbon atoms. The geometries of the pairs were optimized using the HF/6–31G** level of theory within the Cs symmetry. Some of the structures adopted non–planar configurations. Stabilization energies of the planar base pairs were computed at the MP2/6–31G** level of theory. The range of stabilization energies of the studied pairs varied between – 24 to – 9 kcal/mol, and the available experimental values agreed well with the calculated gas phase interaction enthalpies within 2 kcal/mol. Analysis of the MP2 stabilization energies showed that the Hartree–Fock energy controlled the stabilization of the base pairs. This means that the electrostatic interactions are responsible for their stability. The correlation interaction energy for weakly bonded base pairs was found to contribute 30–40% to the stabilization, and for some base pairs repulsive correlation interaction energy was even observed [76].
The structure and stability of nucleic acids were estimated through hydrogen–bonded nucleic acid base pairs dominated by electrostatic interactions. The RI–MP2 (resolution of identity MP2) method in combination with the cc–pVTZ basis set was used for geometry optimizations. The MP2 optimizations usually led to the improvement of the intermolecular energy terms due to intramolecular geometry adjustments such as the N–H bond elongation in base pairs. The interaction energies were calculated using the Complete Basis Set (CBS) extrapolation within the Dunning’s basis set series. These energies ranged from –5 kcal/mol for the weakest non–polar pairs to –47 kcal/mol for the strongest protonated ones. Coupled–Cluster corrections with inclusion of non–iterative triple contributions (CCSD(T)) was used for some base pairs. To guarantee the accuracy of 0.5 to 1.0 kcal/mol for these H–bonded base pairs the MP2 extrapolation to the CBS limit must be based on the aug–cc–pVQZ basis set. In addition, the ΔCCSD(T) correction must also be included. The PW91 DFT functional with the 6–31G** basis set reproduced the RI–MP2/CBS interaction energies with a maximum error of 2.6 kcal/ mol, when applied on the B3LYP optimized geometries. Interaction energies calculated with the B3LYP functional deviated from the RI– MP2/CSB results by with a few kcal/mol, with a relative error of 2.2 kcal/mol [77]. It is important to remember that the DFT methods (in contrast to the MP2 method) are usually not suitable for base stacking calculations described above.
Recent studies of the oligonucleotide crystals showed that the base pair interactions seemed to be stabilized by non–planar DNA base amino groups. Therefore, an exact description of the geometry and deformability of the DNA base amino groups is very important. The MP2/6–31–G* level of theory was used to compare planar and non–planar geometries of adenine, cytosine, guanine, thymine, and isocytosine. The range of dihedral angles between the cytosine and adenine rings and their amino group hydrogen atoms was found to lie between 10 to 25°, and the angles between the two guanine amino group hydrogen atoms and the guanine ring were 43 and 12°. The non–planar structures of cytosine and adenine were only 0.4 kcal/ mol more stable than the planar ones, whereas the non–planar guanine was 1.6 kcal/mol more stable than the planar molecule [78]. In summary, the amino group non–planarity was more pronounced for guanine than for adenine and cytosine, with the larger basis sets favouring planar structures. For example, preliminary calculations of formamide [79] and formamidine [80] illustrated a decrease in the amino group non–planarity on going to extended basis sets (6– 311G(3df,2p) and 5s4p3df/3s2p). In the case of formamidine the energy difference between planar and non–planar structures decreased from 0.85 to 0.3 kcal/mol (in comparison with MP2/6– 31G*), while the amino group hydrogen atom dihedrals were reduced by less than 15%. The higher theoretical level used for formamide [79,80] yielded a planar structure and non–planarity of this system was extremely small with any basis set [79, 80]. As a result, larger basis sets are required for geometry optimisations.
The most recent study by P. Hobza et al. [81] showed that the best and most well balanced description of non–covalent interactions in DNA base pairs was produced with the MP2/cc–pVTZ method. For cyclic hydrogen bonds such as those found in nucleic acid base pairs the MP2 method paired with any basis set did not yield reliable results except a very large basis set, aug–cc–pVTZ. It has to be stressed that cyclic hydrogen bonds are not particularly common within proteins or most protein–ligand complexes, and so this deficiency of the MP2 method is not important for studies of DNA base pairs. It was noted that normally MP2 interaction energies were quite poorly described when the counterpoise correction method was not employed. The lowest average unsigned interaction energy errors with values of 0.67 and 0.70 kcal/mol were obtained in combination with the aug–cc– pVTZ and cc–pVTZ basis sets, respectively. Excellent accuracy was achieved for hydrogen bonding and mixed interactions with the aug–cc–pVTZ basis set, whereas the cc–pVTZ basis set produced better results for each of the interaction types and described mixed interactions particularly well. Among smaller basis sets, 6– 31G*(0.25) and TZVP produced the best results. Both of these basis sets yielded an average error of 1.32 kcal/mol with large errors for hydrogen bonding interactions and relatively low ones for dispersion interactions. In cases where the size of large system does not allow to use large basis sets with the MP2 method and only qualitative results are required, the 6–31G* basis was recommended as the best choice to study non–covalent interaction energies.
A combination of quantum chemical calculations and molecular dynamics simulations was used to study the role of a solvent on tautomerism of nucleic acid bases and structure and properties of nucleic acid base pairs [82]. Particular attention was paid to micro–hydrated (by one or two water molecules) complexes, whose structures found by scanning empirical potential surfaces were re–calculated at a correlated ab initio level of theory. Additionally, isolated as well as mono– and di–hydrated H–bonded, T–shaped and stacked structures of all possible nucleic acid base pairs were studied at the same level of theory. Water as solvent was shown to have a strong influence on the equilibrium between the tautomers of bases as well as the spatial arrangement of the bases in a base pair. The results provided clear evidence that the prevalence of either the stacked or hydrogen–bonded structures of the base pairs in the solvent was not determined only by their bulk properties but rather by specific hydrophilic interactions of the base pair with a small number of solvent molecules [82]. For example, the canonical form of cytosine became favored in the presence of just two water molecules accompanied by the most favorable free energy of hydration. For guanine unusual tautomers with very large dipole moments, albeit extremely disfavored energetically in the gas phase (by about 20 kcal mol-1), were significantly stabilized by water. The hydration of adenine also reduced the difference between the stability of the canonical form (global minimum) and the first two local minima, which resulted in the co–existence of these three forms. The canonical forms of thymine and uracil were undoubtedly favored both in the gas phase and in the water environment. These trends were also confirmed by calculations of the hydration free energies. The following conclusions were drawn in the study:
(1) Only marginal differences were found between the calculated hydration energies of tautomers of bases using any of the following models: conductor–like polarizable continuum model (C–PCM), molecular dynamics thermal integration (MD–TI) method and hybrid methods.
(2) The largest discrepancy between the hydration energies was found for tautomers with high dipole moments, whose results should therefore be interpreted with care. The higher the hydration number of the base pairs, the lower the difference between the stability of hydrogen bonded (HB) complexes and single (S) molecule structures. This means that S structures hydrated better than the HB ones as a result of a denser network of H–bonds than in the case of HB structures. The presence of two water molecules brought additional 6 kcal momol-1 to the stability of S structures in comparison with the HB base pair, regardless of their chemical structure.
(3) Methylation of bases led to an even stronger preference for stacked structures, which are favored over the H–bonded ones even in the absence of water molecules in the majority of cases. Preference for stacked structures in the DNA base pairs in the water solution might arise from the hydrophilic interaction of a small number of water molecules and not only, as expected, from the hydrophobic effect of bulk water [82].
Effect of Specific Interactions on Vibrational Frequencies
The preference of normal tautomeric forms of nucleobases in aqueous solution was demonstrated by Raman measurements [83]. This result was consistent with the pKA values of adenine and its tautomer, adenine* that differ by a factor of 105 [84]. The important role of tautomers of nucleobases in mutagenesis has been studied with experimental and theoretical methods [85]. Quantum chemical calculations of vibrational frequencies of both forms can guide identification of rare tautomers in solution or in gas phase [86]. In the case of the adenine–thymine (AT) complex and the complex between corresponding imino–enol tautomers, adenine*–thymine* (A*T*), geometry optimizations at the SCF/MINI–1 level of theory indicated that the former complex was more stable by about 10 kcal/ mol than the latter. Single–point HF calculations with a larger basis set, MIDI–1, on the optimized geometries optimized resulted in unrealistic interaction energies. Therefore, single–point calculations of this kind should be taken with great care for the AT and A*T* complexes. Out of the two semi– empirical methods, AM1 and PM3, the PM3 results for the geometries, harmonic force fields, heats of formation, interaction energies and the energy difference between the AT and A*T* complexes were in better agreement with the SCF/ MINI–1 results. The SCF/MINI–1 interaction enthalpy of –11.32 kcal/mol for the AT complex matched well the experimental value of –12.9 kcal/mol. The deformation energy and basis set superposition error were found to be of importance in these complexes. Knowledge of the harmonic vibrational spectra of isolated A, T, A*, and T* bases and the AT and A*T* base pairs made it possible to determine the frequency and intensity changes induced by the Watson–Crick type of hydrogen bonding in the AT base pair and tautomeric transitions between the AT and A*T* complexes [86]. In the AT base pair, the vibrational spectra of A and T were still separate, thus making the frequency shifts and intensity changes due to base pairing predictable in experimental vibrational spectra. This was not found to be true for the A*T* complex due to significant coupling of the normal vibrations between A* and T*, which resulted in strong in–plane ring vibrational modes of isolated bases and N–H bending modes of adenine upon formation of hydrogen bonds. The appearance of new intensive bands related to the stretching modes of the N3=C4 and C6=N10 double bonds of T* and A*, respectively, in the spectral regions was suggested as a clear marker for the spectroscopic detection of the existence of these rare tautomers in solutions (Figure 7).
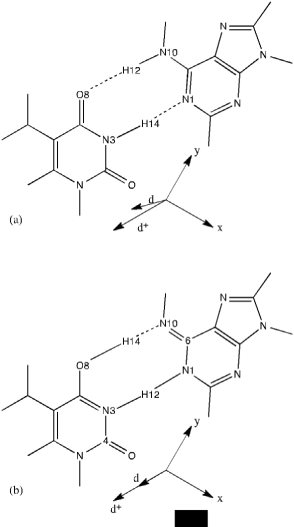
Figure 7: SCF/MINI–1 optimized structures and dipole moments d+ of
isolated (a) adenine*–thymine* and (b) adenine–thymine complexes [86].
Polarization–dependent two–dimensional infrared (2D IR) spectra of the purine and pyrimadine base vibrations of five nucleotide monophosphates (NMPs) were obtained in D2O at neutral pH in the frequency range 1500–1700 cm-1. In contrast with the traditional peak assignment, the distinctive cross-peaks between the ring deformations and carbonyl stretches of NMPs indicated that these vibrational modes were highly coupled. The traditional peak assignment is based on a simple local mode picture such as C=O, C=N, and C=C double bond stretches. Except in the case of guanine and cytosine that have a single local CO stretching mode, no simple or intuitive structural correlations were found to readily assign the spectral features. Density functional theory (DFT) calculations with explicit waters solvating hydrogen–bonding sites showed that multiple ring vibrations were indeed coupled and delocalized over the purine and pyrimidine rings. Generally, there was a good agreement between the experimental and computational results, thus forming building blocks for constructing a structure– based model of DNA and RNA vibrational spectroscopy [87].
The results of harmonic and an harmonic frequency calculations on a guanine–cytosine complex with an enolic structure (a tautomeric form with cytosine in the enol form and with a hydrogen at the 7– position on guanine) were compared with gas–phase IR–UV double resonance spectral data. Harmonic frequencies were obtained at the RI–MP2/cc–pVDZ, RI–MP2/TZVPP, and semi–empirical PM3 levels of electronic structure theory. An–harmonic frequencies were obtained by the correlation–consistent vibrational self–consistent field (CC–VSCF) method with improved PM3 potential surfaces. The average absolute percentage deviations were 2.6% for harmonic RI– MP2/cc–pVDZ (3.0% with a scaling factor of 0.956 to compensate for anharmonicity), 2.5% for harmonic RI–MP2/TZVPP (2.9% with a scaling factor of 0.956 to account for anharmonicity), and 2.3% for adapted PM3 CC–VSCF. Inclusion of the empirical scaling factor in the ab initio harmonic calculations usually improves the stretching frequencies but decreases the accuracy of the other mode frequencies. The anharmonic calculations indicated that anharmonicity along single mode coordinates could be significant for simple stretching modes. For several cases, coupling between different vibrational modes provided the main contribution to anharmonicity. Examples of strongly an–harmonically coupled modes included the symmetric stretch and group torsion of the hydrogen–bonded NH2 group on guanine, the OH stretch and torsion of the enol group on cytosine, and the NH stretch and the NH out–of–plane bend of non– hydrogen bonded NH group on guanine [88]. The CC–VSCF method with adapted PM3 potential surfaces was found to be useful in obtaining accurate frequency information for biological molecules. Further improvements in the PM3 method, such as in describing hydrogen bonds [89,90], might further improve the accuracy. Most importantly, the power of gas–phase spectroscopy provided accurate information on increasingly complex biological molecules. Additional biological systems can therefore be explored by the combination of high– resolution spectroscopy and harmonic and anharmonic vibrational calculations with high–quality potentials. This should lead to more understanding about the properties of the potential surfaces of biological molecules [88].
Intermolecular vibrational modes of the H–bonded adenine... thymine Watson–Crick (ATWC) base pair were studied using multidimensional non–harmonic treatment. Relying on a Born– Oppenheimer–like separation of the fast and slow vibrational motions, the complete multi–dimensional vibrational problem was reduced to a six–dimensional sub–problem, in which all rearrangements between the pair fragments (i.e. adenine and thymine) could be described. The potential energy surface is determined at the Hartree– Fock level with minimal basis set (HF/MINI–1) and an analytical potential energy function was obtained by fitting to the ab initio data. This function was used to calculate vibrational energy levels and effective geometries within the framework of the Hamiltonian model, disregarding the role of the kinematic and potential (in–plane)–(out– of–plane) interactions. The results were in reasonable agreement with the normal coordinate analysis (harmonic treatment); thus indicating physical precision of this standard approach for an approximate description of the lowest vibrational states of the AT base pair. In addition, harmonic vibrational frequencies of the AT pair and 28 other base pairs were evaluated at the HF/MINI–1 and HF/6–31G** levels of theory. Both methods produced deviation in intermolecular harmonic vibrational frequencies by less than 30%. For all the base pairs, the buckle and propeller vibrational modes [for definition and nomenclature see R. E. Dickerson et al., EMBO J. 8, 1 (1989)] were the lowest ones, all being in the narrow interval (from 4 to 30 cm-1 in the harmonic approximation). Although most of the evaluated harmonic frequencies were qualitative, in some cases due to a strong non– harmonic behavior of the buckle motion, the calculated vibrations became physically meaningless. For all the base pairs the buckle and propeller vibrations were the lowest vibrational modes. While the buckle motion was shown to be harmonic for some pairs (adenine– thyamine and guanine–cytosine) it was found to be strongly anharmonic for the other ones such as guanine–guanine complex. Other approaches need to be used for quantitative description of these anharmonic modes [91,92].
The subtle conformational changes that occur upon tautomeric equilibria are usually difficult to understand, which renders the spectral assignment a complex task. To understand the structural and spectroscopic properties of this kind of systems a full vibrational spectroscopic study of the 7H–keto– amino tautomeric form of guanine was performed, (both for the isolated molecule and the condensed phase). By the combination of all the available spectroscopic vibrational techniques (FTIR, Raman, and inelastic neutron spectroscopy (INS)) with state–of–the–art theoretical approaches such as DFT, a complete and exact assignment of the experimental spectra was achieved. For studying of nucleic acid bases, condensed–phase periodic DFT calculations were used in this study [93]. Between predicted and experimental spectra, a very good agreement was achieved for the Raman and INS data (both regarding frequencies and intensities). Particularly regarding the INS profile, detailed features such as Davydov splittings and vibrational modes related to intermolecular H–bond interactions could be unequivocally assigned for the first time. The study also highlighted that periodic functionals with the Plane–Wave basis sets was needed to predict vibrational frequencies of the molecule in the solid state. In particular, the low energy region of the spectrum, including external (lattice) modes, could only be accurately predicted through such a plane–wave methodology [93].
Podolyan et al. [94] reports gas–phase B3LYP, HF, and MP2 calculations of the vibrational infrared spectra of the two DNA base pairs such as adenine–thymine and guanine–cytosine. A few basis sets, including 6–31G(d), 6–31++G(d,p), aug–cc–pVDZ, and cc–pVTZ, were used at the HF and B3LYP levels of theory to assess vibrational modes and their IR intensities. Although it was not possible to compare calculated frequencies to experimental data (due to the lack of thereof in gas phase), the comparative performance of these methods showed that all these levels of theory produced similar results. Therefore, an appropriate DFT functional with a medium– sized basis set is adequate for theoretical evaluation of the IR spectra of studies complexes. Therefore, the use of a more expensive electron–correlated method such as MP2 was not justified. At the same time, out of these methods only the MP2 method identified the non–planarity of the some DNA base pairs [94–124].
In summary, it appears that electron correlated methods such as MP2 and CCSD(T) are important in providing a balanced description of intermolecular interactions in DNA base pairs. The DFT field has recently exploded with a number of new DFT functionals that are supposed to better treat medium– and long–range correlation effects. Inclusion of an explicit correction to treat dispersion interactions has been shown to significantly improve their performance. Therefore, a more comprehensive study on the performance of wave function– and DFT–based methods is needed to select the best combination of a quantum chemical method and basis set for studying intermolecular interactions and IR frequencies/intensities of oligonucleotides in gas phase and in solution, thus paving the way towards a more comprehensive description of structure and properties of DNA. It appears that the solvent effects on conformational changes (especially those related to non–planarity) cannot be neglected and solvent molecules need to be explicitly included in calculations to help elucidate factors that are responsible for the unique hydration effect in DNA [125–180].
Future Studies
(1) We are particularly interested in explaining why the asymmetric phosphodiester mode of nucleic acids shifts from 1225 cm-1 in the B–DNA conformation to 1240 cm-1 in the A–DNA form for double stranded DNA and single stranded DNA but not RNA. We are also interested in understanding why the intensity of the symmetric stretch at 1080 cm-1 in B–DNA is much greater than that in A–DNA.
(2) Predict vibrational frequencies and IR intensities of single nucleotides and oligonucleotides by means of methods of computational quantum chemistry, including those of density functional theory and correlated levels of theory such as MP2 and CCSD(T). The following factors that affect the accurate prediction of vibrational frequencies will be systematically studied: (a) conformation of the nucleotide/oligonucleotide and the effect of intramolecular hydrogen bonding and pi–pi stacking, (b) effect of basis set, and (c) solvent effects: implicit through a polarisable continuum model or explicit on vibrational frequencies and intensities, etc. This goal will be achieved by comparing the experimental FTIR spectra with the calculated frequencies and intensities. As a result, a new computational methodology for accurate prediction of vibrational frequencies of oligonucleotides will be established.
(3) Explain why the asymmetric phosphodiester mode of nucleic acids shifts from 1225 cm-1 in the B– DNA conformation to 1240 cm-1 in the A–DNA form in response to dehydration for double and single standard DNA. We are also interested in why the intensity of the symmetric stretch at 1080 cm-1 in B– DNA is much greater than the intensity of this mode in A–DNA.
References
- Heidari, A Thesis submitted to the Faculty of the Chemistry, California South University (CSU), Irvine, California, The United States of America (USA) in Fulfillment of the Requirements for the Degree of Doctor of Philosophy (PhD) in Chemistry, Academic Year 2012.
- DNA.
- Russell P. iGenetics. New York: Benjamin Cummings. 2001; ISBN 0-8053-4553-1.
- Dahm R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum Genet. 2008; 122: 565-581.
- Jones ME. Albrecht Kossel, a biographical sketch. Yale J Biol Med. 1953; 26: 80-97.
- Levene P. The structure of yeast nucleic acid. J Biol Chem. 1919; 40: 415–424.
- Astbury WT. X-ray studies of nucleic acids. Symp Soc Exp Biol. 1947; : 66-76.
- Soyfer VN. The consequences of political dictatorship for Russian science. Nat Rev Genet. 2001; 2: 723-729.
- Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994; 58: 563-602.
- Avery O, MacLeod C, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944; 79: 137–158.
- Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952; 36: 39-56.
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953; 171: 737-738.
- The B-DNA X-ray pattern on the right of this linked image was obtained by Rosalind Franklin and Raymond Gosling in May 1952 at high hydration levels of DNA and it has been labeled as "Photo 51".
- Nature Archives Double Helix of DNA: 50 Years.
- Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953; 171: 740-741.
- Original X-ray diffraction image. Osulibrary.oregonstate.edu. 2011.
- Wilkins MH, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953; 171: 738-740.
- The Nobel Prize in Physiology or Medicine 1962.
- Maddox B. The double helix and the 'wronged heroine'. Nature. 2003; 421: 407-408.
- Crick FHC (Lecture, 1955, Retrieved 2006) On degenerate templates and the adaptor hypothesis.
- Meselson M, Stahl FW. The replication of DNA in Escherichia Coli. Proc Natl Acad Sci U S A. 1958; 44: 671-682.
- The Nobel Prize in Physiology or Medicine 1968.
- Mandelkern M, Elias JG, Eden D, Crothers DM. The dimensions of DNA in solution. J Mol Biol. 1981; 152: 153-161.
- Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, Dunham A, et al. The DNA sequence and biological annotation of human chromosome 1. Nature. 2006; 441: 315-321.
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953; 171: 737-738.
- Berg J, Tymoczko J, Stryer L. Biochemistry. W. H. Freeman and Company. 2002.
- Abbreviations and Symbols for Nucleic Acids, Polynucleotides and their Constituents IUPAC-IUB Commission on Biochemical Nomenclature (CBN). 2006.
- Ghosh A, Bansal M. A glossary of DNA structures from A to Z. Acta Crystallogr D Biol Crystallogr. 2003; 59: 620-626.
- Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006; 34: 564-574.
- Molecular structure of the b-dna dodecamer d(cgcaaatttgcg)2; an examination of propeller twist and minor-groove water structure at 2.2 angstroms resolution. 1993. PDB 1D65.
- Basu HS, Feuerstein BG, Zarling DA, Shafer RH, Marton LJ. Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies. J Biomol Struct Dyn. 1988; 6: 299-309.
- Franklin RE, Gosling RG. The structure of sodium thymonucleate fibers I. The influence of water content. Acta Crystallogr. 1953; 6: 673–677; Franklin RE, Gosling RG. The structure of sodium thymonucleate fibres. II. The cylindrically symmetrical Patterson function. Acta Crystallogr. 6: 678–685.
- Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL. Polymorphism of DNA double helices. J Mol Biol. 1980; 143: 49-72.
- Baianu IC. Structural order and partial disorder in biological systems. Bull. Math. Biol. 1980; 42: 137–141.
- Hosemann R, Bagchi RN. Direct analysis of diffraction by matter, North-Holland Publs., Amsterdam – New York. 1962; 734.
- Baianu IC. X-ray scattering by partially disordered membrane systems. Acta Crystallogr A. 1978; 34: 751–753.
- Wahl MC, Sundaralingam M. Crystal structures of A-DNA duplexes. Biopolymers. 1997; 44: 45-63.
- Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J Mol Biol. 2000; 300: 819-840.
- Rothenburg S, Koch-Nolte F, Haag F. DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles. Immunol Rev. 2001; 184: 286-298.
- Oh DB, Kim YG, Rich A. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc Natl Acad Sci U S A. 2002; 99: 16666-16671.
- Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003; 4: 566-572.
- Fraser DB. The Interpretation of Infrared Dichroism in Fibrous Protein Structures. J. Chem. Phys. 1953; 21: 1511.
- Rill RL. The linear dichroism of oriented helical and superhelical polymers. Biopolymers. 1972; 11: 1929-1941.
- Brahms J, Pilet J, Phuong Lan TT, Hill LR. Direct evidence of the C-like form of sodium deoxyribonucleate. Proc Natl Acad Sci U S A. 1973; 70: 3352-3355.
- Sutherland GBBM, Tsuboi M. The Infra-Red Spectrum and Molecular Configuration of Sodium Deoxyribonucleate. Proc. Roy. Soc (London). 1957; A239: 446.
- Tsuboi M. Vibrational Spectra of Phosphite and Hypophosphite Anions, and the Characteristic Frequencies of PO3-- and PO2- Groups. J. Am. Chem. Soc. 1957; 79: 1351.
- Shimanouchi T, Tsuboi M, Kyogoku Y. The structure and properties of biomolecules and biological systems. Advances in Chemical Physics,. New York: Interscience. 1964; 7: 435-437.
- Derrick MR, Humecki H J: Editor. Infrared microspectroscopy in the analysis of cultural artifacts’, in Practical guide to infrared microspectroscopy. New York: Marcel Dekker. 287–322.
- Newman R. Some applications of infrared spectroscopy in the examination of painting materials. J. Am. Institute Conservation. 1980; 19: 42–62.
- Feller RL: Editor Artists’ pigments: A handbook of their history and characteristics, Vol. 1.Cambridge, UK: Cambridge University Press. 1986.
- Roy A: Editor Artists’ pigments: A handbook of their history and characteristics, Vol. 2. Washington, DC, USA: National Gallery of Art. 1993.
- West-Fitzhugh E: Editor Artists’ Pigments: A Handbook of Their History and Characteristics, Vol. 3. Washington, DC, USA: National Gallery of Art. 1997.
- Visser T, Meyers RA: Editor Infrared spectroscopy in environmental analysis’, in Encyclopedia of Analytical Chemistry, Vol. 1. Chichester, UK: Wiley. 2000; 1–21.
- Spencer HM. International critical tables, Vol. I. New York: McGraw-Hill. 1926; 67.
- Ojha RP, Dhingra MM, Sarma MH, Shibata M, Farrar M, Turner CJ, Sarma RH . DNA bending and sequence-dependent backbone conformation NMR and computer experiments. Eur J Biochem. 1999; 265: 35-53.
- Feughelman M, Langridge R, Seeds WE, Stokes AR, Wilson HR, Hooper CW, et al. Molecular structure of deoxyribose nucleic acid and nucleoprotein. Nature. 1955; 175: 834-838.
- Bradbury EM, Price WC, Wilkinson GR. Infrared studies of molecular configurations of DNA. J. Mol. Biol. 1961; 3: 301.
- Ramachandran KI, Deepa G, Namboori K. Computational chemistry and molecular modeling: Principles and Applications, Springer-Verlag, Berlin Heidelberg. 2008.
- Koch W, Holthausen MC. A chemist’s guide to density functional theory, Second Edition, WILEY-VCH Verlag GmbH, D-69469 Weinheim, Federal Republic of Germany. 2001.
- Cramer CJ. Essentials of computational chemistry: Theories and Models, Second Edition, John Wiley & Sons Ltd, The Atrium, Southern Gate, Chichester, West Sussex PO19 8SQ, England. 2004.
- Jensen F. Introduction to computational chemistry, Second Edition, John Wiley & Sons Ltd The Atrium, Southern Gate, Chichester, West Sussex PO19 8SQ, England. 2007.
- Muller-Dethlefs K, Hobza P. Noncovalent Interactions: A Challenge for Experiment and Theory. Chem. Rev. 2000; 100: 143–167.
- Vondrásek J, Bendová L, Klusák V, Hobza P. Unexpectedly strong energy stabilization inside the hydrophobic core of small protein rubredoxin mediated by aromatic residues: correlated ab initio quantum chemical calculations. J Am Chem Soc. 2005; 127: 2615-2619.
- Tsuzuki S, Honda K, Uchimaru T, Mikami M. Ab initio calculations of structures and interaction energies of toluene dimers including CCSD(T) level electron correlation correction. J Chem Phys. 2005; 122: 144323.
- Sinnokrot MO, Sherrill CD. Substituent effects in pi-pi interactions: sandwich and T-shaped configurations. J Am Chem Soc. 2004; 126: 7690-7697.
- Tsuzuki S, Honda K, Uchimaru T, Mikami M, Tanabe K. The Magnitude of the CH/π Interaction between Benzene and Some Model Hydrocarbons. J. Am. Chem. Soc. 2000; 122: 3746–3753.
- Hobza P, Sponer J. Structure, energetics, and dynamics of the nucleic Acid base pairs: nonempirical ab initio calculations. Chem Rev. 1999; 99: 3247-3276.
- Pittner J, Hobza P. CCSDT and CCSD(T) calculations on model H-bonded and stacked complexes. Chem. Phys. Lett. 2004; 390: 496–499.
- Pérez A, Sponer J, Jurecka P, Hobza P, Luque FJ, Orozco M. Are the hydrogen bonds of RNA (AU) stronger than those of DNA (AT)? A quantum mechanics study. Chemistry. 2005; 11: 5062-5066.
- Jurecka P, Hobza P. True stabilization energies for the optimal planar hydrogen-bonded and stacked structures of guanine...cytosine, adenine...thymine, and their 9- and 1-methyl derivatives: complete basis set calculations at the MP2 and CCSD(T) levels and comparison with experiment. J Am Chem Soc. 2003; 125: 15608-15613.
- Sponer J, Jurecka P, Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J Am Chem Soc. 2004; 126: 10142-10151.
- Jurecka P, Šponer J, Hobza P. Potential Energy Surface of the Cytosine Dimer: MP2 Complete Basis Set Limit Interaction Energies, CCSD(T) Correction Term, and Comparison with the AMBER Force Field. J. Phys. Chem. B. 2004; 108: 5466–5471.
- Dabkowska I, Gonzalez HV, Jurecka P, Hobza P. Stabilization energies of the hydrogen-bonded and stacked structures of nucleic acid base pairs in the crystal geometries of CG, AT, and AC DNA steps and in the NMR geometry of the 5'-d(GCGAAGC)-3' hairpin: Complete basis set calculations at the MP2 and CCSD(T) levels. J Phys Chem A. 2005; 109: 1131-1136.
- Hobza P, Sponer J. Toward true DNA base-stacking energies: MP2, CCSD(T), and complete basis set calculations. J Am Chem Soc. 2002; 124: 11802-11808.
- Jurecka P, Sponer J, Cerný J, Hobza P. Benchmark database of accurate (MP2 and CCSD(T) complete basis set limit) interaction energies of small model complexes, DNA base pairs, and amino acid pairs. Phys Chem Chem Phys. 2006; 8: 1985-1993.
- Šponer J, Leszczynski J, Hobza P. Structures and energies of hydrogen-bonded DNA base pairs; A nonempirical study with inclusion of electron correlation. J. Phys. Chem. 1996; 100: 1965-1974.
- Sponer J, Jurecka P, Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J Am Chem Soc. 2004; 126: 10142-10151.
- Šponer J, Hobza P. Nonplanar geometries of DNA bases: Ab Initio second-order Møller-Plesset study. J. Phys. 1994; 98: 3161-3164.
- Kwiatkowski JS, Leszczynski JJ. Molecular structure and vibrational IR spectrum of formamide revisited: ab initio post-Hartree—Fock study. Mol. Sruct. 1993; 297: 277.
- Šponer J, Hobza P, Unpublished results.
- Riley KE, Hobza P. Assessment of the MP2 method, along with several basis sets, for the computation of interaction energies of biologically relevant hydrogen bonded and dispersion bound complexes. J Phys Chem A. 2007; 111: 8257-8263.
- Kabelác M, Hobza P. Hydration and stability of nucleic acid bases and base pairs. Phys Chem Chem Phys. 2007; 9: 903-917.
- Hartman KA, Lord RC, Thomas Jr. GJ, Duchesne J: Editor. In physiccchemical properties of nucleic acids, Academic Press, London, Vol. 2. 1973.
- Wolfenden RV. Tautomeric equilibria in inosine and adenosine. J Mol Biol. 1969; 40: 307-310.
- Kwiatkowski JS, Person WB, Editors: Bcveridge DL, Lavery R. Theoretical biochemistry and molecular biophysics, Adenine Press, Guilderland, New York. 1990; 1: p 153.
- Hrouda V, Florián J, Hobza P. Structure, energetics, and harmonic vibrational spectra of the adenine-thymine and adenine*-thymine* base pairs: Gradient nonempirical and semiempirical study. J. Phys. Chem. 1993; 97: 1542-1551.
- Peng CS, Jones KC, Tokmakoff A. Anharmonic vibrational modes of nucleic acid bases revealed by 2D IR spectroscopy. J Am Chem Soc. 2011; 133: 15650-15660.
- Brauer B, Gerber RB, Kabelác M, Hobza P, Bakker JM, Abo Riziq AG, etal. Vibrational spectroscopy of the G…C base pair: Experiment, harmonic and anharmonic calculations, and the nature of the anharmonic couplings. J. Phys. Chem. 2005; A 109: 6974-6984.
- Harb W, Bernal-Uruchurtu MI, Ruiz-Lo´pez, MF. An improved semiempirical method for hydrated systems. Theor. Chem. Acc. 2004; 112: 204-216.
- Dannenberg JJ. Hydrogen bonds: a comparison of semiempirical and ab initio treatments. THEOCHEM. 1997; 401: 279-286.
- Špirko V, Šponer J, Hobza P. Anharmonic and harmonic intermolecular vibrational modes of the DNA base pairs. J. Chem. Phys. 1997; 106: 1472-.1479.
- We did not use scaling since our aim was to compare harmonic and non-harmonic approaches. We are aware of the fact that scaling removes overestimation of vibrational frequencies (harmonic as well as non-harmonic) originating from the Hartree–Fock description.
- Lopes RP, Marques MPM, Valero R, Tomkinson J, Batista de Carvalho LAE. Guanine: A combined study using vibrational spectroscopy and theoretical methods. Spectroscopy: An International Journal. 2012; 27: 5-6: 273–292.
- Podolyana Y, Nowakb MJ, Lapinskib L, Leszczynski J. Probing ab initio MP2 approach towards the prediction of vibrational infrared spectra of DNA base pairs. Journal of Molecular Structure. 2005; 744–747: 19–34.
- D Lacerenza, S Aneli, M Omedei, S Gino, S Pasino, P Berchialla, et al. A molecular exploration of human DNA/RNA co-extracted from the palmar surface of the hands and fingers. Forensic Science International: Genetics. 2016; 22: 44-53.
- Wenjing Lu, Jidong Wang, Qiong Wu, Jiashu Sun, Yiping Chen, Lu Zhang, et al. High-throughput sample-to-answer detection of DNA/RNA in crude samples within functionalized micro-pipette tips. Biosensors and Bioelectronics. 2016; 75: 28-33.
- Pramod kumar, Deepak R. Jain. Cy-Aminopropylene peptide nucleic acid (amp-PNA): chiral cationic PNAs with superior PNA: DNA/RNA duplex stability and cellular uptake, Tetrahedron. 2015; 71: 3378-3384.
- Sorci L, Ruggieri S, Raffaelli N. NAD homeostasis in the bacterial response to DNA/RNA damage. DNA Repair (Amst). 2014; 23: 17-26.
- José C Nunes, Maria Teresa Pessoa de Amorim, Isabel C Escobar, João A Queiroz, António M Morão. Plasmid DNA/RNA separation by ultrafiltration: Modeling and application study. Journal of Membrane Science. 2014; 463: 1-10.
- Oliver DK, Maddocks, Christiaan F. Labuschagne, Peter D Adams, Karen H Vousden. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Molecular Cell. 2016; 61: 210-221.
- Prakash C Joshi, Hsin H Li, Monique Merchant, Thomas C Keane. Total inhibition of 1O2-induced oxidative damage to guanine bases of DNA/RNA by turmeric extracts. Biochemical and Biophysical Research Communications. 2014; 452: 515-519.
- Farukh Arjmand, Imtiyaz Yousuf, Taibi ben Hadda, Loic Toupet. Synthesis, crystal structure and antiproliferative activity of Cu(II) nalidixic acid–DACH conjugate: Comparative in vitro DNA/RNA binding profile, cleavage activity and molecular docking studies. European Journal of Medicinal Chemistry. 2014; 81: 76-88.
- Ivana Stolic, Katarina Miškovic, Ivo Piantanida, Mirela Baus Loncar, Ljubica Glavaš-Obrovac, Miroslav Bajic. Synthesis, DNA/RNA affinity and antitumour activity of new aromatic diamidines linked by 3,4-ethylenedioxythiophene. European Journal of Medicinal Chemistry. 2011; 46: 743-755.
- Marijana Radic Stojkovic, Saška Marczi, Ljubica Glavaš-Obrovac, Ivo Piantanida. Permanent positive charge strongly influences DNA/RNA binding and antiproliferative activity of urea–phenanthridinium conjugates. European Journal of Medicinal Chemistry. 2010; 45: 3281-3292.
- Anders Jorgensen, Katrine Maigaard, Gitta Wörtwein, Ida Hageman, Trine Henriksen, Allan Weimann, et al. Corrigendum to: “Chronic restraint stress in rats causes sustained increase in urinary corticosterone excretion without affecting cerebral or systemic oxidatively generated DNA/RNA damage”. Prog Neuropsychopharmacol Biol Psychiatry 40 (2013) 30–37. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2015; 58: 105.
- João Conde, Elazer R. Edelman, Natalie Artzi. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: The jack-of-all-trades in cancer nanotheranostics? Advanced Drug Delivery Reviews. 2015; 81: 169-183.
- Smith S, Jefferies C. Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev. 2014; 25: 745-757.
- Jorgensen A, Maigaard K, Wörtwein G, Hageman I, Henriksen T, Weimann A, et al. Chronic restraint stress in rats causes sustained increase in urinary corticosterone excretion without affecting cerebral or systemic oxidatively generated DNA/RNA damage. Prog Neuropsychopharmacol Biol Psychiatry. 2013; 40: 30-37.
- Jitka Vondrušková, Jaroslav Kypr, Iva Kejnovská, Markéta Fialová, Michaela Vorlícková. Role of loops in the guanine quadruplex formation by DNA/RNA hybrid analogs of G4T4G4. International Journal of Biological Macromolecules. 2008; 43: 463-467.
- Ivana Stolic, Katarina Miškovic, Anahi Magdaleno, Ariel Mariano Silber, Ivo Piantanida, Miroslav Bajic, et al. Effect of 3,4-ethylenedioxy-extension of thiophene core on the DNA/RNA binding properties and biological activity of bisbenzimidazole amidines. Bioorganic & Medicinal Chemistry. 2009; 17: 2544-2554.
- Marijana Radic Stojkovic, Ivo Piantanida. Tuning urea–phenanthridinium conjugates for DNA/RNA and base pair recognition. Tetrahedron. 2008; 64: 7807-7814.
- Brian E Lang, Frederick P Schwarz. Thermodynamic dependence of DNA/DNA and DNA/RNA hybridization reactions on temperature and ionic strength. Biophysical Chemistry. 2007; 131: 96-104.
- Kohji Seio, Munefumi Tokugawa, Hirosuke Tsunoda, Akihiro Ohkubo, Fumio Arisaka, Mitsuo Sekine. Assembly of pyrene-modified DNA/RNA duplexes incorporating a G-rich single strand region. Bioorganic & Medicinal Chemistry Letters. 2013; 23: 6822-6824.
- AJ Privett, JA Morales. Electron nuclear dynamics of proton collisions with DNA/RNA bases at ELab = 80 keV: A contribution to proton cancer therapy research. Chemical Physics Letters. 2014; 603: 82-88.
- Piotr Guga, Malgorzata Boczkowska, Magdalena Janicka, Anna Maciaszek, Slawomir Kuberski, Wojciech J Stec. Unusual Thermal Stability of RNA/[RP-PS]-DNA/RNA Triplexes Containing a Homopurine DNA Strand. Biophysical Journal. 2007; 92: 2507-2515.
- Philippe Cloutier, Mathieu Lavallée-Adam, Denis Faubert, Mathieu Blanchette, Benoit Coulombe. Methylation of the DNA/RNA-binding protein Kin17 by METTL22 affects its association with chromatin. Journal of Proteomics. 2014; 100: 115-124.
- Irena Kratochvílová, Martin Vala, Martin Weiter, Miroslava Špérová, Bohdan Schneider, Ondrej Páv, et al. Charge transfer through DNA/DNA duplexes and DNA/RNA hybrids: Complex theoretical and experimental studies. Biophysical Chemistry. 2013; 180–181: 127-134.
- Shu Yang, Sadaf T Warraich, Garth A Nicholson, Ian P Blair. Fused in sarcoma/translocated in liposarcoma: A multifunctional DNA/RNA binding protein. The International Journal of Biochemistry & Cell Biology. 2010; 42: 1408-1411.
- Liu CC, Yeung CY, Chen PH, Yeh MK, Hou SY. Salmonella detection using 16S ribosomal DNA/RNA probe-gold nanoparticles and lateral flow immunoassay. Food Chem. 2013; 141: 2526-2532.
- Mirna Marinic, Ivo Piantanida, Gordana Rusak, Mladen Žinic. Interactions of quercetin and its lanthane complex with double stranded DNA/RNA and single stranded RNA: Spectrophotometric sensing of poly G. Journal of Inorganic Biochemistry. 2006; 100: 288-298.
- Anders Jorgensen, Jesper Krogh, Kamilla Miskowiak, Tom G Bolwig, Lars V Kessing, Anders Fink-Jensen, et al. Systemic oxidatively generated DNA/RNA damage in clinical depression: Associations to symptom severity and response to electroconvulsive therapy. Journal of Affective Disorders. 2010; 149: 355-362.
- Jagadeesan Lakshmipraba, Sankaralingam Arunachalam, Anvarbatcha Riyasdeen, Rajakumar Dhivya, Sivanandham Vignesh, Mohammad Abdulkader Akbarsha, etaal. DNA/RNA binding and anticancer/antimicrobial activities of polymer–copper(II) complexes. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy. 2013; 109: 23-31.
- Kamil Malác, Ivan Barvík. Complex between Human RNase HI and the phosphonate-DNA/RNA duplex: Molecular dynamics study. Journal of Molecular Graphics and Modelling. 2013; 44: 81-90.
- Thomas Jacroux, Daniel C Rieck, Rong Cui, Yexin Ouyang, Wen-Ji Dong. Enzymatic amplification of DNA/RNA hybrid molecular beacon signaling in nucleic acid detection. Analytical Biochemistry. 2013; 432: 106-114.
- Qi Zhang, Yang Cai, Hui Li, De-Ming Kong, Han-Xi Shen. Sensitive dual DNAzymes-based sensors designed by grafting self-blocked G-quadruplex DNAzymes to the substrates of metal ion-triggered DNA/RNA-cleaving DNAzymes. Biosensors and Bioelectronics. 2012; 38: 331-336.
- Momoko Kitaoka, Hirofumi Ichinose, Masahiro Goto. Simultaneous visual detection of single-nucleotide variations in tuna DNA using DNA/RNA chimeric probes and ribonuclease A. Analytical Biochemistry. 2009; 389: 6-11.
- Yuichiro Kanno, Takafumi Serikawa, Jun Inajima, Yoshio Inouye. DP97, a DEAD box DNA/RNA helicase, is a target gene-selective co-regulator of the constitutive androstane receptor. Biochemical and Biophysical Research Communications. 2012; 426: 38-42.
- Nicole C Robb, Thorben Cordes, Ling Chin Hwang, Kristofer Gryte, Diego Duchi, Timothy D Craggs, et al. The Transcription Bubble of the RNA Polymerase– Promoter Open Complex Exhibits Conformational Heterogeneity and Millisecond-Scale Dynamics: Implications for Transcription Start-Site Selection. Journal of Molecular Biology. 2013; 425: 875-885.
- Mayte Suárez-Fariñas, Benjamin Ungar, Joel Correa da Rosa, David A Ewald, Mariya Rozenblit, Juana Gonzalez, et al. RNA sequencing atopic dermatitis transcriptome profiling provides insights into novel disease mechanisms with potential therapeutic implications. Journal of Allergy and Clinical Immunology. 2015; 135: 1218-1227.
- SG Parisi, L Sarmati, S Andreis, R Scaggiante, M Cruciani, R Ferretto, et al. Strong and persistent correlation between baseline and follow-up HIV-DNA levels and residual viremia in a population of naïve patients with more than 4 years of effective antiretroviral therapy. Clinical Microbiology and Infection. 21: 288. e5-288.e7.
- Johanna Hass, Esther Walton, Carrie Wright, Andreas Beyer, Markus Scholz, Jessica Turner, et al. Associations between DNA methylation and schizophrenia-related intermediate phenotypes — A gene set enrichment analysis. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2015; 59: 31-39.
- Luísa Camacho, Mallikarjuna S Basavarajappa, Ching-Wei Chang, Tao Han, Tetyana Kobets, et al. Effects of oral exposure to bisphenol A on gene expression and global genomic DNA methylation in the prostate. female mammary gland, and uterus of NCTR Sprague-Dawley rats, Food and Chemical Toxicology. 2015; 81: 92-103.
- C Haas, E Hanson, MJ Anjos, R Banemann, A Berti, E Borges, et al. RNA/DNA co-analysis from human saliva and semen stains – Results of a third collaborative EDNAP exercise. Forensic Science International: Genetics. 2013; 7: 230-239.
- Mariane Noronha Domingues, Mauricio Luis Sforça, Adriana Santos Soprano, Jack Lee, Tatiana de Arruda Campos Brasil de Souza, et al. Structure and Mechanism of Dimer–Monomer Transition of a Plant Poly(A)-Binding Protein upon RNA Interaction: Insights into Its Poly(A) Tail Assembly. Journal of Molecular Biology. 2015; 427: 2491-2506.
- C Haas, E Hanson, MJ Anjos, KN Ballantyne, R Banemann, B Bhoelai, et al. RNA/DNA co-analysis from human menstrual blood and vaginal secretion stains: Results of a fourth and fifth collaborative EDNAP exercise. Forensic Science International: Genetics. 2014; 8: 203-212.
- Samir Ounzain, Rudi Micheletti, Carme Arnan, Isabelle Plaisance, Dario Cecchi, Blanche Schroen, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. Journal of Molecular and Cellular Cardiology.2015; 89: 98-112.
- Owen M Peters, Gabriela Toro Cabrera, Helene Tran, Tania F Gendron, Jeanne E McKeon, Jake Metterville, et al. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron. 2015; 88: 902-909.
- Sen SK, Boelte KC, Barb JJ, Joehanes R, Zhao X, Cheng Q, et al. Integrative DNA, RNA, and protein evidence connects TREML4 to coronary artery calcification. Am J Hum Genet. 2014; 95: 66-76.
- Onur Keskin, Heiner Wedemeyer, Ali Tüzün, Kalliopi Zachou, Xheni Deda, George N Dalekos, et al. Association Between Level of Hepatitis D Virus RNA at Week 24 of Pegylated Interferon Therapy and Outcome. Clinical Gastroenterology and Hepatology. 2015; 13: 2342-2349.e2.
- Yoshihiro Matsumura, Ryo Nakaki, Takeshi Inagaki, Ayano Yoshida, Yuka Kano, Hiroshi Kimura, et al. H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Molecular Cell. 2015; 60: 584-596.
- Wei Gan, Ben Nie, Fei Shi, Xin-Min Xu, Jian-Chang Qian, Yasumitsu Takagi, et al. Age-dependent increases in the oxidative damage of DNA, RNA, and their metabolites in normal and senescence-accelerated mice analyzed by LC–MS/MS: Urinary 8-oxoguanosine as a novel biomarker of aging. Free Radical Biology and Medicine. 2012; 52: 1700-1707.
- William H Chappell, Stephen L Abrams, Kvin Lertpiriyapong, Timothy L Fitzgerald, Alberto M Martelli, Lucio Cocco, et al. Novel roles of androgen receptor, epidermal growth factor receptor, TP53, regulatory RNAs, NF-kappa-B, chromosomal translocations, neutrophil associated gelatinase and matrix metalloproteinase-9 in prostate cancer and prostate cancer stem cells. Advances in Biological Regulation. 2016; 60: 64-87.
- Elena Alexandrova, Nicola Miglino, Adnan Hashim, Giovanni Nassa, Claudia Stellato, Michael Tamm, et al. Small RNA profiling reveals deregulated phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/Akt pathway in bronchial smooth muscle cells from asthmatic patients. Journal of Allergy and Clinical Immunology. 2016; 137: 58-67.
- Zhong-Fa Zhang, Daisuke Matsuda, Sok Kean Khoo, Kristin Buzzitta, Elizabeth Block, David Petillo, et al. A comparison study reveals important features of agreement and disagreement between summarized DNA and RNA data obtained from renal cell carcinoma. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2008; 657: 77-83.
- Bernadette M Boac, Yin Xiong, Douglas C Marchion, Forough Abbasi, Stephen H Bush, Ingrid J Ramirez, et al. Micro-RNAs associated with the evolution of ovarian cancer cisplatin resistance. Gynecologic Oncology. 2016; 140: 259-263.
- Jaakko LO Pohjoismäki, J Bradley Holmes, Stuart R Wood, Ming-Yao Yang, Takehiro Yasukawa, Aurelio Reyes, et al. Mammalian Mitochondrial DNA Replication Intermediates Are Essentially Duplex but Contain Extensive Tracts of RNA/DNA Hybrid. Journal of Molecular Biology. 2010; 397: 1144-1155.
- Wenjing Wu, Tushar D Bhagat, Xue Yang, Jee Hoon Song, Yulan Cheng, Rachana Agarwal, et al. Hypomethylation of Noncoding DNA Regions and Overexpression of the Long Noncoding RNA, AFAP1-AS1, in Barrett's Esophagus and Esophageal Adenocarcinoma. Gastroenterology. 2013; 144: 956-966.e4.
- Simona Incollu, Maria Barbara Lepori, Antonietta Zappu, Valentina Dessì, Maria Cristina Noli, Eva Mameli, et al. DNA and RNA studies for molecular characterization of a gross deletion detected in homozygosity in the NH2-terminal region of the ATP7B gene in a Wilson disease patient. Molecular and Cellular Probes. 2011; 25: 195-198.
- Shile Zhang, Jun S Wei, Samuel Q Li, Tom C Badgett, Young K Song, Saurabh Agarwal, et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Letters. 2016; 371: 214-224.
- King-Hwa Ling, Peter J Brautigan, Sarah Moore, Rachel Fraser, Pike-See Cheah, Joy M Raison, et al. Derivation of an endogenous small RNA from double-stranded Sox4 sense and natural antisense transcripts in the mouse brain. Genomics. 2016; 107: 88-99.
- C Haas, E Hanson, MJ Anjos, W Bär, R Banemann, A Berti, et al. RNA/DNA co-analysis from blood stains—Results of a second collaborative EDNAP exercise. Forensic Science International: Genetics. 2012; 6: 70-80.
- Farukh Arjmand, Imtiyaz Yousuf, Mohd Afzal, Loic Toupet. Design and synthesis of new Zn(II) nalidixic acid– DACH based Topo-II inhibiting molecular entity: Chemotherapeutic potential validated by its in vitro binding profile, pBR322 cleavage activity and molecular docking studies with DNA and RNA molecular targets. Inorganica Chimica Acta. 2014; 421: 26-37.
- Jean-Michel Cayuela, Carole Mauté, Anne-Lise Fabre, Olivier Nibourel, Stéphanie Dulucq, Eric Delabesse, et al. A novel method for room temperature distribution and conservation of RNA and DNA reference materials for guaranteeing performance of molecular diagnostics in onco-hematology: A GBMHM study. Clinical Biochemistry. 2015; 48: 982-987.
- Sébastien Lyonnais, Robert J Gorelick, Fatima Heniche-Boukhalfa, Serge Bouaziz, Vincent Parissi, Jean-François Mouscadet, et al. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: From nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Research. 2013; 171: 287-303.
- Leila Sabrina Ullmann, Claudia de Camargo Tozato, Camila Dantas Malossi, Tais Fukuta da Cruz, Raíssa Vasconcelos Cavalcante, Jacqueline Kazue Kurissio, et al. Comparative clinical sample preparation of DNA and RNA viral nucleic acids for a commercial deep sequencing system (Illumina MiSeq®). Journal of Virological Methods. 2015; 220: 60-63.
- Kasai K, Nishiyama N, Izumi Y, Otsuka S, Ishihara A, Yamauchi K. Exposure to 3,3',5-triiodothyronine affects histone and RNA polymerase II modifications, but not DNA methylation status, in the regulatory region of the Xenopus laevis thyroid hormone receptor βΑ gene. Biochem Biophys Res Commun. 2015; 467: 33-38.
- C Haas, E Hanson, R Banemann, AM Bento, A Berti, Á Carracedo, et al. RNA/DNA co-analysis from human skin and contact traces – results of a sixth collaborative EDNAP exercise. Forensic Science International: Genetics. 2015; 16: 139-147.
- Michael O Clarke, Richard Mackman, Daniel Byun, Hon Hui, Ona Barauskas, Gabriel Birkus, et al. Discovery of ß-d-2'-deoxy-2'-a-fluoro-4'-a-cyano-5-aza-7,9-dideaza adenosine as a potent nucleoside inhibitor of respiratory syncytial virus with excellent selectivity over mitochondrial RNA and DNA polymerases. Bioorganic & Medicinal Chemistry Letters. 2015; 25: 2484-2487.
- Giorgia Botta, Bruno Mattia Bizzarri, Adriana Garozzo, Rossella Timpanaro, Benedetta Bisignano, Donatella Amatore, et al. Carbon nanotubes supported tyrosinase in the synthesis of lipophilic hydroxytyrosol and dihydrocaffeoyl catechols with antiviral activity against DNA and RNA viruses. Bioorganic & Medicinal Chemistry. 2015; 23: 5345-5351.
- Kirill A Afonin, Eckart Bindewald, Maria Kireeva, Bruce A Shapiro. Chapter Thirteen - Computational and Experimental Studies of Reassociating RNA/DNA Hybrids Containing Split Functionalities, In: Shi-Jie Chen and Donald H Burke-Aguero, Editor(s), Methods in Enzymology. Academic Press. 2015; 553: 313-334.
- Vikash Singh, Gajendra Kumar Azad, Santhosh Kumar Sariki, Raghuvir S Tomar. Flocculation in Saccharomyces cerevisiae is regulated by RNA/DNA helicase Sen1p. FEBS Letters. 2015; 589: 3165-3174.
- Matthew J Schellenberg, Percy P Tumbale, R Scott Williams. Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease. Progress in Biophysics and Molecular Biology. 2015; 117: 157-165.
- R Jucker, A Sulzer, P Voegeli, C Haas, A Kratzer. In-house validation of an RNA/DNA co-extraction strategy for body fluid identification/STR profiling and application to forensic casework. Forensic Science International: Genetics Supplement Series. 2015; 5: e381-e383.
- Tri-Nhan Nguyen, Dong-Ju You, Hiroyuki Matsumoto, Eiko Kanaya, Yuichi Koga, Shigenori Kanaya. Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid. Journal of Structural Biology. 2013; 182: 144-154.
- Hiroyuki Ida, Kosuke Fukuda, Akira Tachibana, Toshizumi Tanabe. Long DNA passenger strand highly improves the activity of RNA/DNA hybrid siRNAs. Journal of Bioscience and Bioengineering. 2014; 117: 401-406.
- Yuuki Fujiwara, Katsunori Hase, Keiji Wada, Tomohiro Kabuta. An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochemical and Biophysical Research Communications. 2015; 460: 281-286.
- Dexian Dong, Junyan Li, Qian Gao, Xianqing Huang, Yuquan Xu, Rongxiu Li. Utilizing RNA/DNA hybridization to directly quantify mRNA levels in microbial fermentation samples. Journal of Microbiological Methods. 2009; 79: 205-210.
- Parimal Routh, Pratap Mukherjee, Arnab Dawn, Arun K. Nandi. Self assembly of poly(o-methoxy aniline) with RNA and RNA/DNA hybrids: Physical properties and conformational change of poly(o-methoxy aniline). Biophysical Chemistry. 2009; 143: 145-153.
- My-Nuong Vo, George Barany, Ioulia Rouzina, Karin Musier-Forsyth. HIV-1 Nucleocapsid Protein Switches the Pathway of Transactivation Response Element RNA/DNA Annealing from Loop–Loop “Kissing” to “Zipper”. Journal of Molecular Biology. 2009; 386: 789-801.
- Xipeng Liu, Yang Zhang, Rubing Liang, Jingli Hou, Jianhua Liu. Characterization of the 3' exonuclease of Chlamydophila pneumoniae endonuclease IV on double-stranded DNA and the RNA strand of RNA/DNA hybrid. Biochemical and Biophysical Research Communications. 2007; 361: 987-993.
- Yosuke Tanaka, Keisuke Satoh, Harumi Yamada, Takayuki Takebe, Hideki Nikaido, Satoshi Shiozawa. Assessment of the nutritional status of field-caught larval Pacific bluefin tuna by RNA/DNA ratio based on a starvation experiment of hatchery-reared fish. Journal of Experimental Marine Biology and Ecology. 2008; 354: 56-64.
- Marcin Nowotny, Sergei A Gaidamakov, Rodolfo Ghirlando, Susana M Cerritelli, Robert J Crouch, Wei Yang. Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Molecular Cell. 2007; 28: 264-276.
- Érica AG Vidal, Paul DiMarco, Phillip Lee. Effects of starvation and recovery on the survival, growth and RNA/DNA ratio in loliginid squid paralarvae. Aquaculture. 2006; 260: 94-105.
- Renee Mercaldo-Allen, Catherine Kuropat, Elaine M Caldarone. A model to estimate growth in young-of-the-year tautog, Tautoga onitis, based on RNA/DNA ratio and seawater temperature. Journal of Experimental Marine Biology and Ecology. 2006; 329: 187-195.
- Teodoro Ramirez, Dolores Cortés, Alberto Garcia, Angel Carpena. Seasonal variations of RNA/DNA ratios and growth rates of the Alboran Sea sardine larvae (Sardina pilchardus). Fisheries Research. 2004; 68: 57-65.
- Mary L Kopka, Laurence Lavelle, Gye Won Han, Ho-Leung Ng, Richard E Dickerson. An Unusual Sugar Conformation in the Structure of an RNA/DNA Decamer of the Polypurine Tract May Affect Recognition by RNase H. Journal of Molecular Biology. 2003; 334: 653-665.
- Woo-Seok Gwak, Yosuke Tanaka, Osamu Tominaga, Tatsuo Tsusaki, Masaru Tanaka. Field evaluation by RNA/DNA ratios on post-release nutritional status of released and wild Japanese flounder Paralichthys olivaceus juveniles. Journal of Experimental Marine Biology and Ecology. 2003; 293: 107-124.
- WS Gwak, T Tsusaki, M Tanaka. Nutritional condition, as evaluated by RNA/DNA ratios, of hatchery-reared Japanese flounder from hatch to release. Aquaculture. 2003; 219: 503-514.
- Aaron M Shapiro, Lutfiya Miller-Pinsler, Peter G Wells. Breast cancer 1 (BRCA1)-deficient embryos develop normally but are more susceptible to ethanol-initiated DNA damage and embryopathies. Redox Biology. 2016; 7: 30-38.
- Cheng-Che E Lan, Shu-Mei Huang, Ching-Shuang Wu, Chin-Han Wu, Gwo-Shing Chen. High-glucose environment increased thrombospondin-1 expression in keratinocytes via DNA hypomethylation. Translational Research. 2016; 169: 91-101.e3.